Binding molecules capable of neutralizing rabies virus and uses thereof
a technology of rabies virus and binding molecules, applied in the field of biotechnology and medicine, can solve the problems of insufficient amount, inability to neutralize rabies virus, and inability to meet the needs of patients,
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
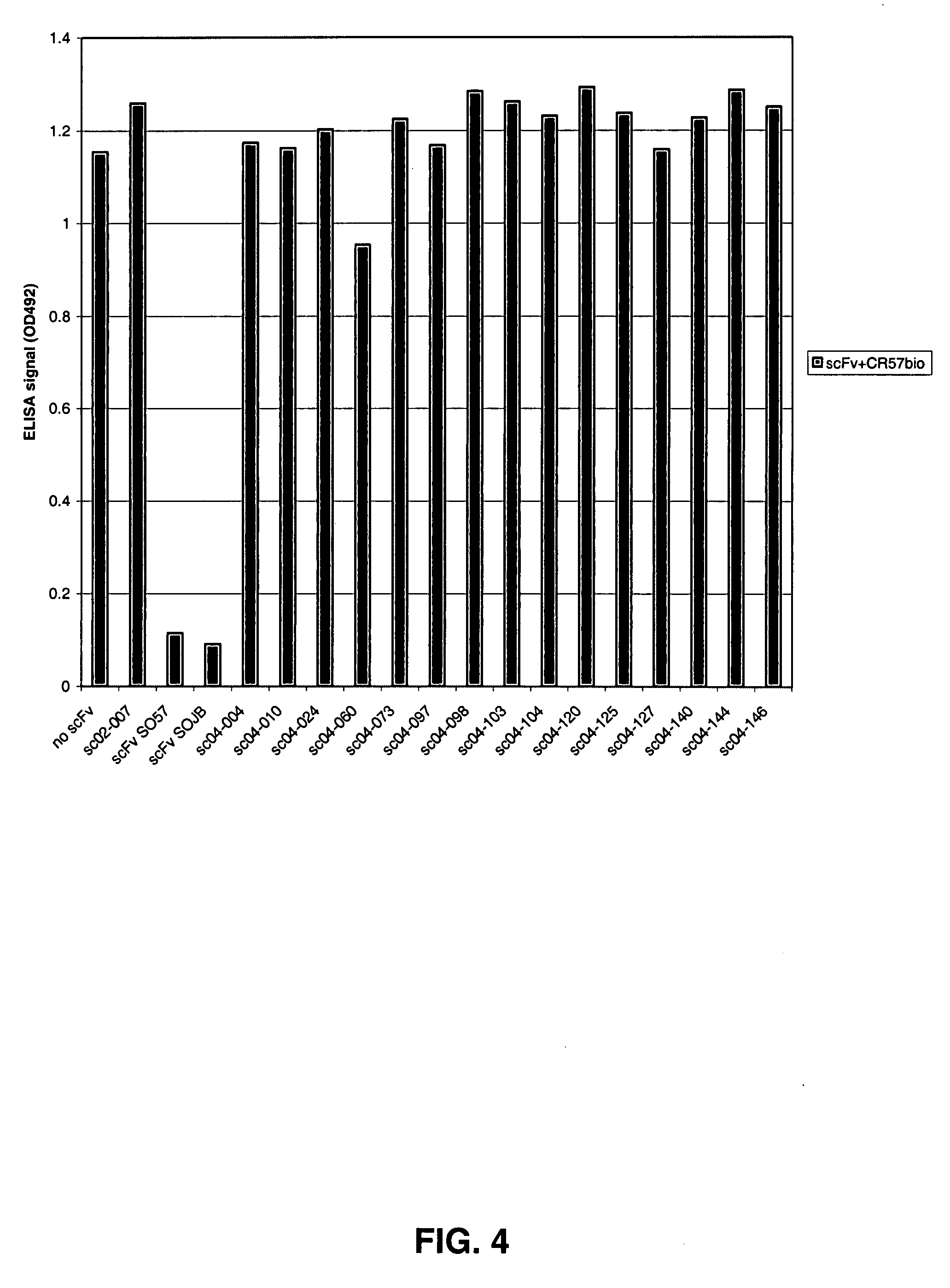
Epitope Recognition of Human Anti-Rabies Antibodies CR-57 and CR-J
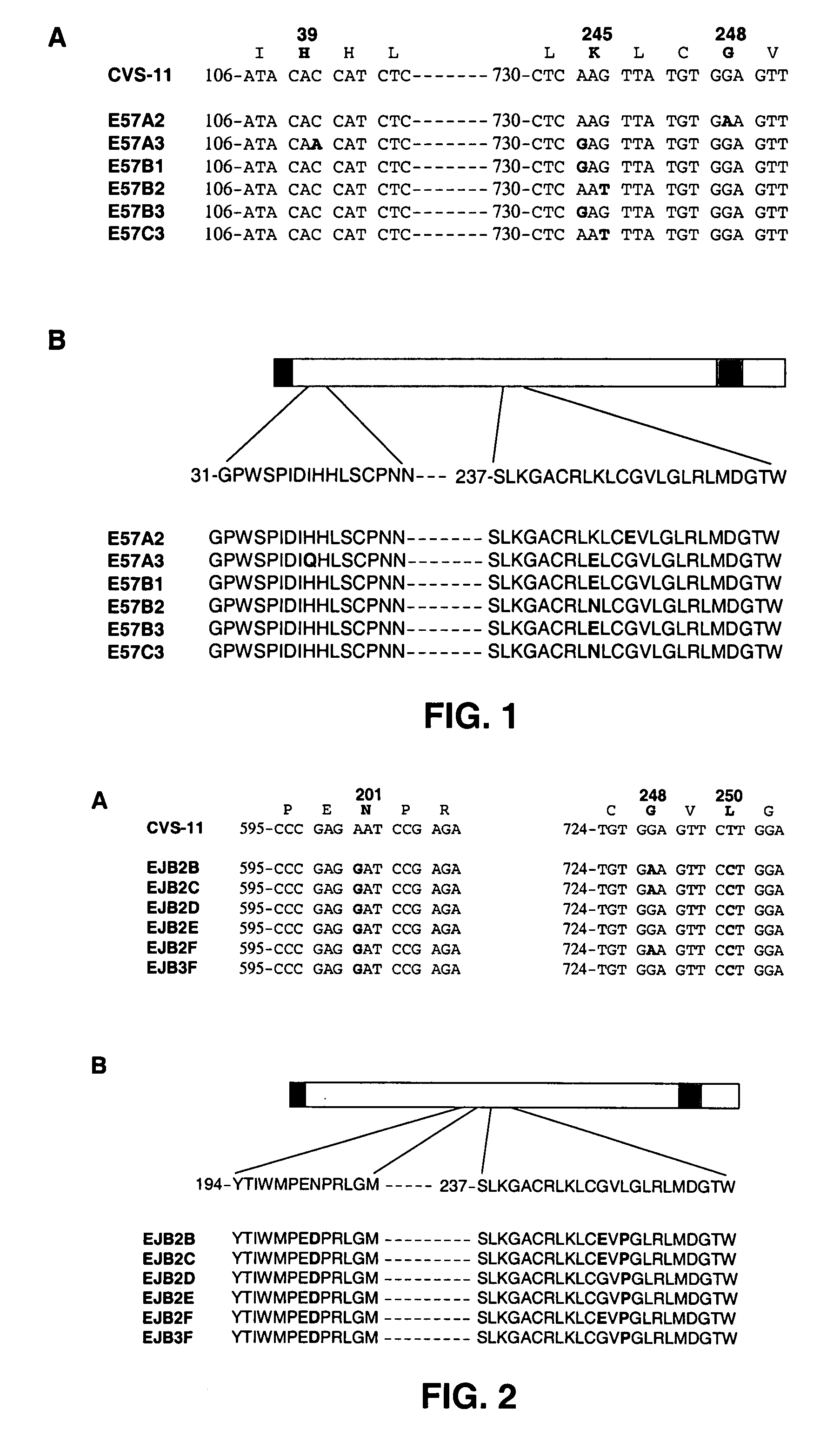
[0121] To address whether the human monoclonal antibodies called CR-57 and CR-JB recognize non-overlapping, non-competing epitopes, escape viruses of the human monoclonal antibodies called CR-57 and CR-JB were generated. CR-57 and CR-JB were generated essentially as described (see, Jones et al., 2003), via introduction of the variable heavy and light chain coding regions of the corresponding antibody genes into a single human IgG1 expression vector named pcDNA3002(Neo). The resulting vectors pgSO57C11 and pgSOJBC11 were used for transient expression in cells from the cell line deposited at the European Collection of Cell Cultures (ECACC), CAMR, Salisbury, Wiltshire SP4 OJG, Great Britain on 29 Feb. 1996 under number 96022940 and marketed under the trademark PER.C6®. The nucleotide and amino acid sequences of the heavy and light chains of these antibodies are shown in SEQ ID NOS:122 through 129, respectively. Serial d...
example 2
Construction of a ScFv Phage Display Library Using Peripheral Blood Lymphocytes of Rabies-Vaccinated Donors
[0126] From four rabies-vaccinated human subjects, 50 ml blood was drawn from a vein one week after the last boost. Peripheral blood lymphocytes (PBL) were isolated from these blood samples using Ficoll cell density fractionation. The blood serum was saved and frozen at −20° C. The presence of anti-rabies antibodies in the sera was tested positive using a FACS staining on rabies virus glycoprotein transfected 293T cells. Total RNA was prepared from the PBL using organic phase separation (TRIZOL™) and subsequent ethanol precipitation. The obtained RNA was dissolved in DEPC-treated ultrapure water and the concentration was determined by OD 260 nm measurement. Thereafter, the RNA was diluted to a concentration of 100 ng / μl. Next, 1 μg of RNA was converted into cDNA as follows: To 10 μl total RNA, 13 μl DEPC-treated ultrapure water and 1 μl random hexamers (500 ng / μl) were added a...
example 3
Selection of Phages Carrying Single Chain Fv Fragments Specifically Recognizing Rabies Virus Glycoprotein
[0131] Antibody fragments were selected using antibody phage display libraries, general phage display technology and MAbstract® technology, essentially as described in U.S. Pat. No. 6,265,150 and in WO 98 / 15833 (both of which are incorporated by reference herein). The antibody phage libraries used were two different semi-synthetic scFv phage libraries (JK1994 and WT2000) and the immune scFv phage libraries (RAB-03-G01 and RAB-04-G01) prepared as described in Example 2 above. The first semi-synthetic scFv phage library (JK1994) has been described in de Kruif et al. (1995b), the second one (WT2000) was built essentially as described in de Kruif et al. (1995b). Briefly, the library has a semi-synthetic format whereby variation was incorporated in the heavy and light chain V genes using degenerated oligonucleotides that incorporate variation within CDR regions. Only VH3 heavy chain ...
PUM
| Property | Measurement | Unit |
|---|---|---|
| volume | aaaaa | aaaaa |
| volume | aaaaa | aaaaa |
| concentration | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com