Identification of bacterial species and subspecies using lipids
a technology of lipids and bacterial species, applied in the field of identification of bacterial species, can solve the problems of reducing the number of false negatives of bacterial species, indirect identification methods have the disadvantage of identifying other bacteria instead, and fewer infectious diseases that can be treated directly by physicians, etc., and achieve the effect of reducing the number of false negatives
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
Identification of Related Bacterial Species
Burkholderia pseudomallei Versus B. thailandensis and B. mallei
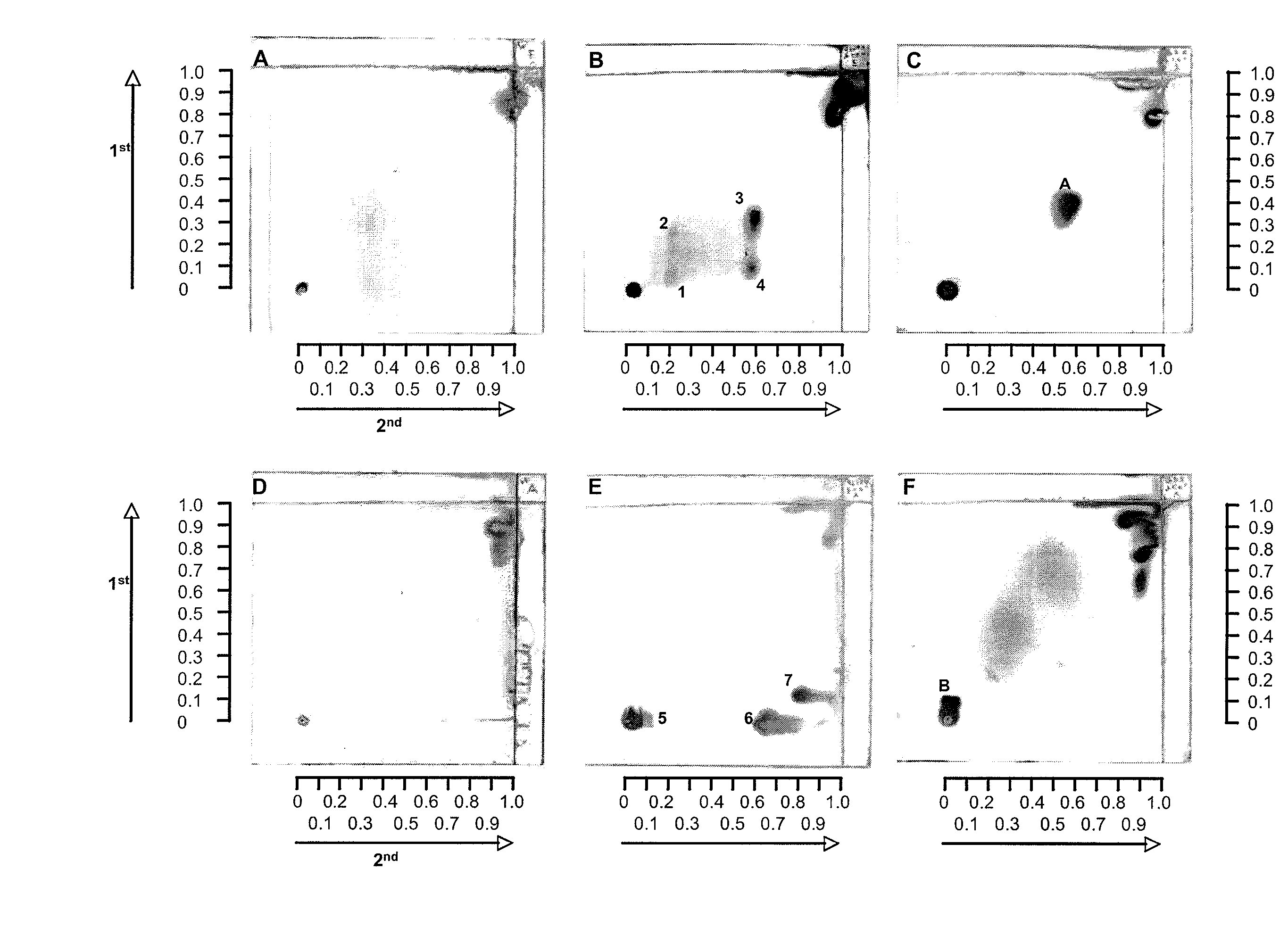
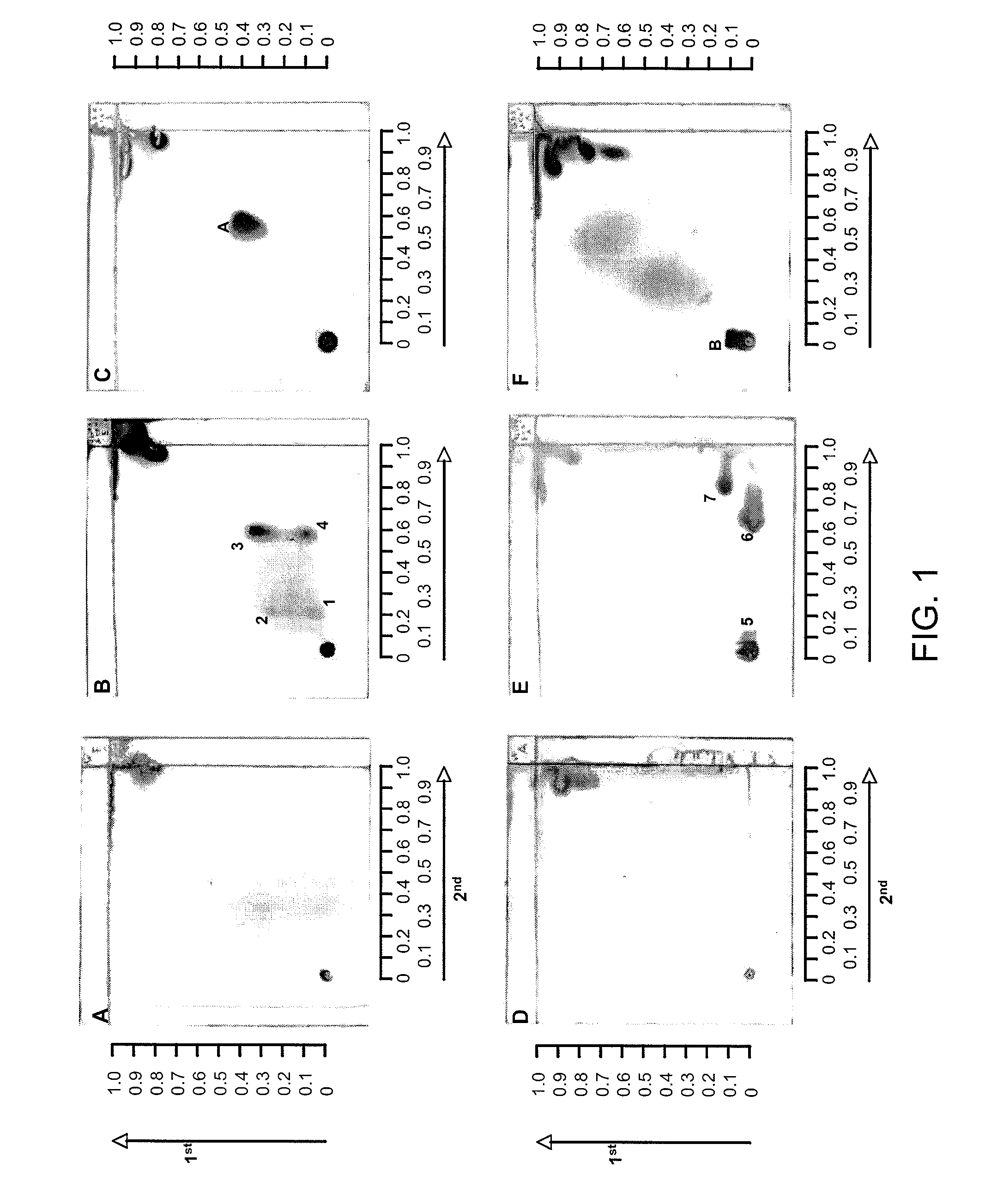
[0042]Burkholderia pseudomallei is the causative agent of melioidosis, a chronic disease in humans. The disease manifests itself in two distinct forms: an acute infection or as an acute bloodstream infection, and as a chronic infection. In chronic or recurrent melioidosis, the lungs are most commonly affected. Mortality is high—up to 86%. This disease is very common in South-East Asia and Northern Australia. A closely related non-pathogenic bacterium, Burkholderia thailandensis, is commonly found in the environment. Another related bacterial pathogen is Burkholderia mallei, the causative agent of glanders, mainly a disease in horses, but can also affect humans. The standard method for laboratory diagnosis of melioidosis is the isolation of the pathogen. This generally takes at least 3 days. Humans with severe infections, especially those with septicemia, often die before resul...
example 2
Identification of Related Bacterial Subspecies (Mycobacterium avium Subspecies Paratuberculosis (MAP) Versus M. avium subsp. avium (MAA) and M. avium subsp. hominissuis (MAH)) by Lipid Profiling and by Mass Spectroscopy
A. Lipid Profiling:
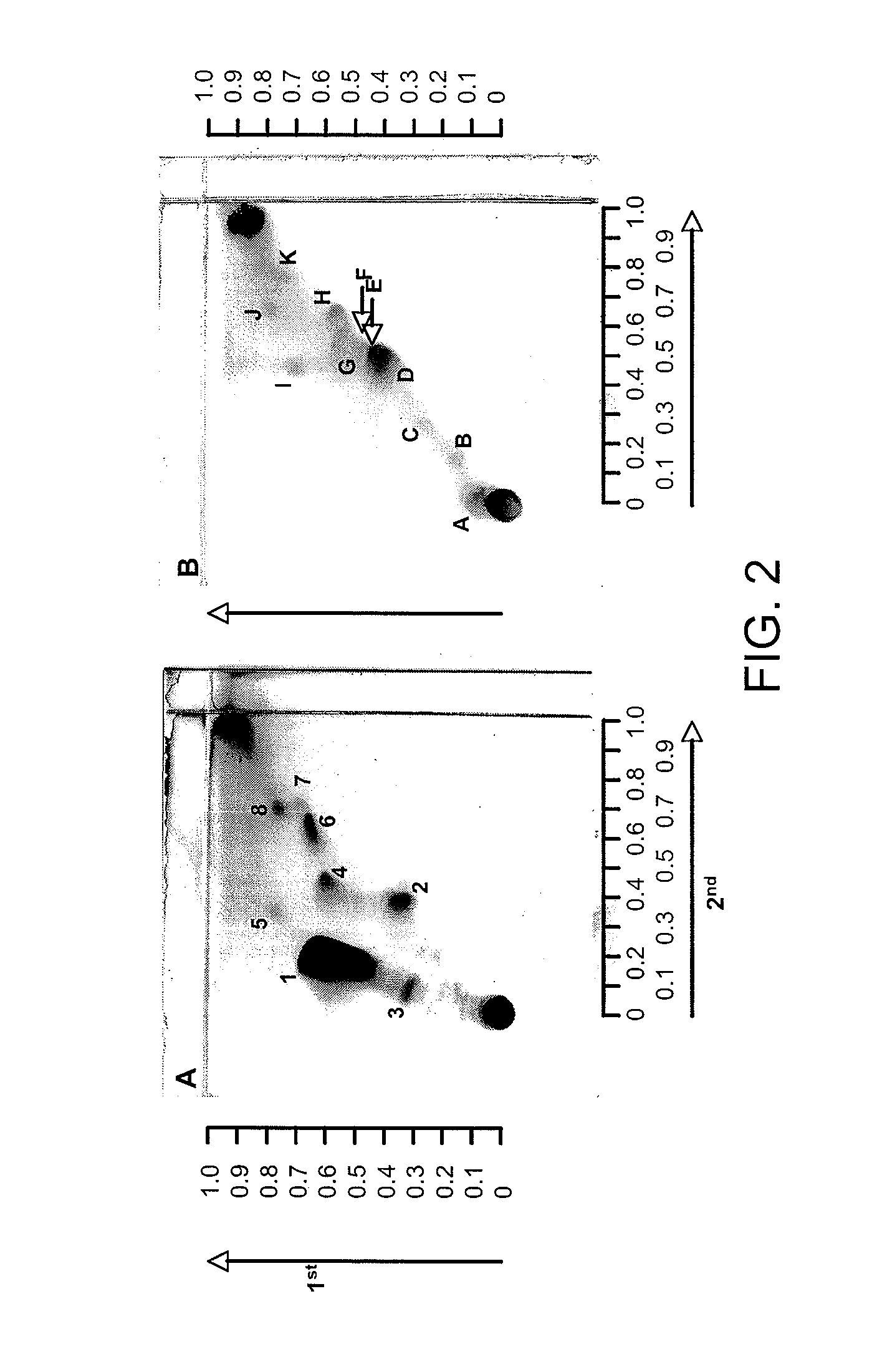
[0050]Mycobacterium avium subspecies paratuberculosis is the causative agent of Johne's disease, a chronic enteritis in ruminants (cattle, sheep), while M. avium subsp. hominissuis and M. avium subsp. avium are closely related subspecies of the same species M. avium that are often found in the environment.
[0051]M. avium subsp. avium (MAA) and M. avium subsp. hominissuis (MAH) are environmental opportunistic pathogen that cause respiratory diseases in the general population which can be distinguished only by the presence or absence of IS901. Environmental mycobacteria are ubiquitous in municipal as well as natural waters, and the primary source of human infection is thought to be water. MAP can be distinguished from MAA / MAH by the presence of IS9...
example 3
Host Immune Response to Mycobacterial Lipids
A. Microtiter Plate Elisa:
[0067] Immunogenic species, such as members of the M. avium complex, exhibit seroreactivity of the glycopeptidolipids, which are immunogenic molecules in the outer part of the cell envelope. Structurally, they consist of a lipopeptide core made out of a tetrapeptide (three amino acids and one amino alcohol) that is N-linked to a mono- or di-unsaturated long fatty acid. O-linked to this core molecule are mono- and oligosaccharides, which may be further modified (for example, by methylation or acylation). These sugar moieties are responsible for the 28 different serovars within this complex. One characteristic of MAP is the absence of those highly immunogenic glycopeptidolipids due to gene decay. Several Studies have focused on the seroreactivities of lipid moieties of mycobacteria, but also lipoglycans (lipoarabinomannan, lipomannan) and oligosaccharides. The seroreactivities of single lipid molecules: phenolic ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com