Microrna modulators and method for identifying and using the same
a technology of microrna and modulator, applied in the field of microrna modulator and method for identifying and using the same, can solve problems such as the effective delivery of such molecules, and achieve the effect of increasing or decreasing the expression of reporter proteins
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
miRNA Inhibitor Screening Assay
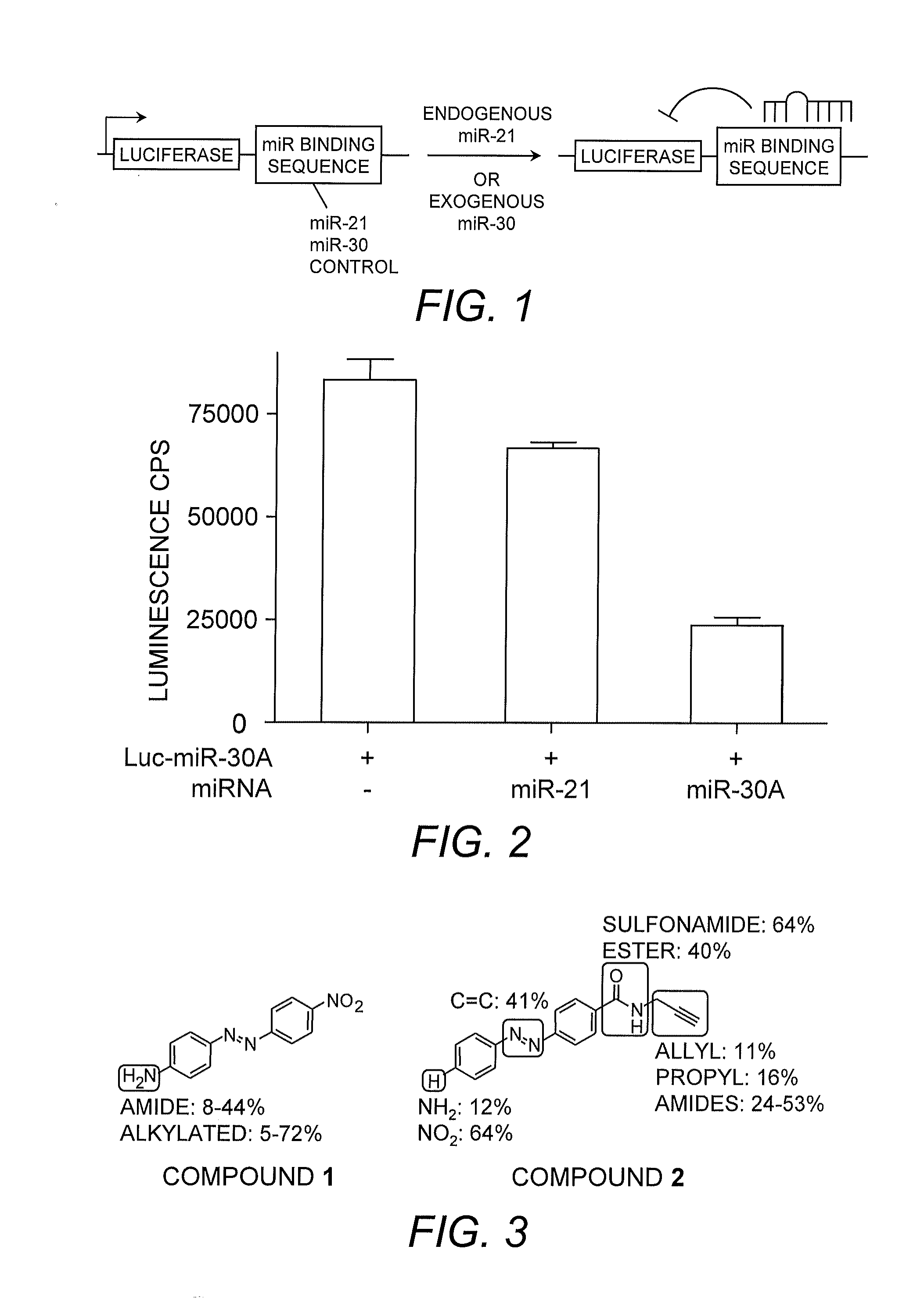
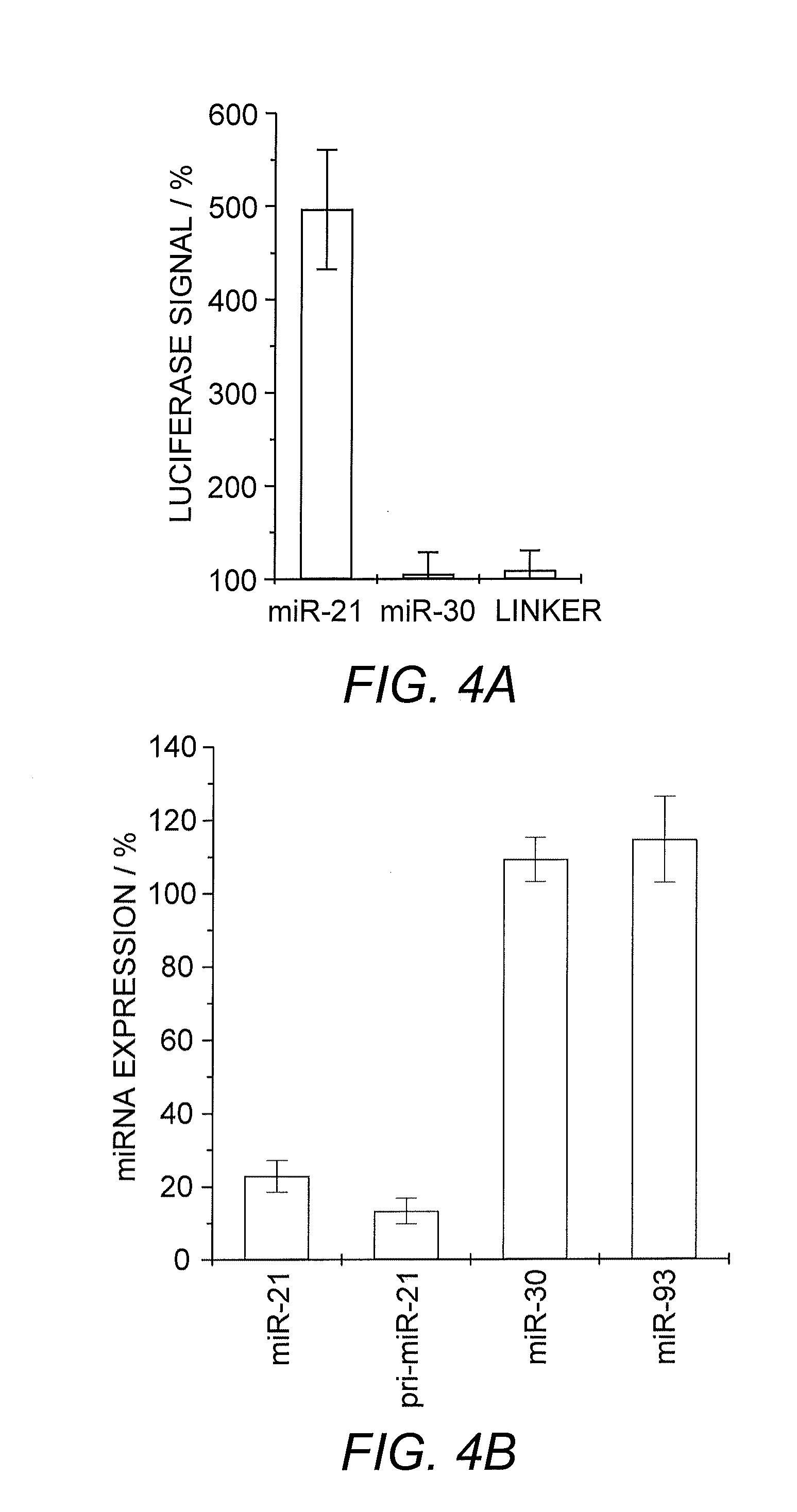
[0054]MicroRNA miR-21 was selected as a target miRNA due to its involvement as an anti-apoptotic factor in cancer cells and its elevated levels in various cancers such as breast, ovarian, and lung cancer as well as glioblastoma (Chan, et al. (2005) Cancer Res. 65:6029). Lentiviral reporter constructs for miRNA activity were assembled by introducing the complementary sequences of mature miR-21 (5′-UAG CUU AUC AGA CUG AUG UUG A-3′; SEQ ID NO:13), miR-30A (5′-CUU UCA GUC GGA UGU UUG CAG C-3′; SEQ ID NO:14) as a specificity control, and a linker sequence (a previously present multiple-cloning site with no detectable recognition by natural miRNAs) downstream of a luciferase reporter gene as a negative control (FIG. 1).
[0055]Luc-miR-21, Luc-miR-30A, and Luc-linker (control) were, by viral infection, introduced into HeLa cells, which express high levels of miR-21, but only low levels of miR-30A (Cheng, et al. (2005) Nucl. Acids Res. 33:1290).
[0056]The specifi...
example 2
Identification of miR-21 Antagonists
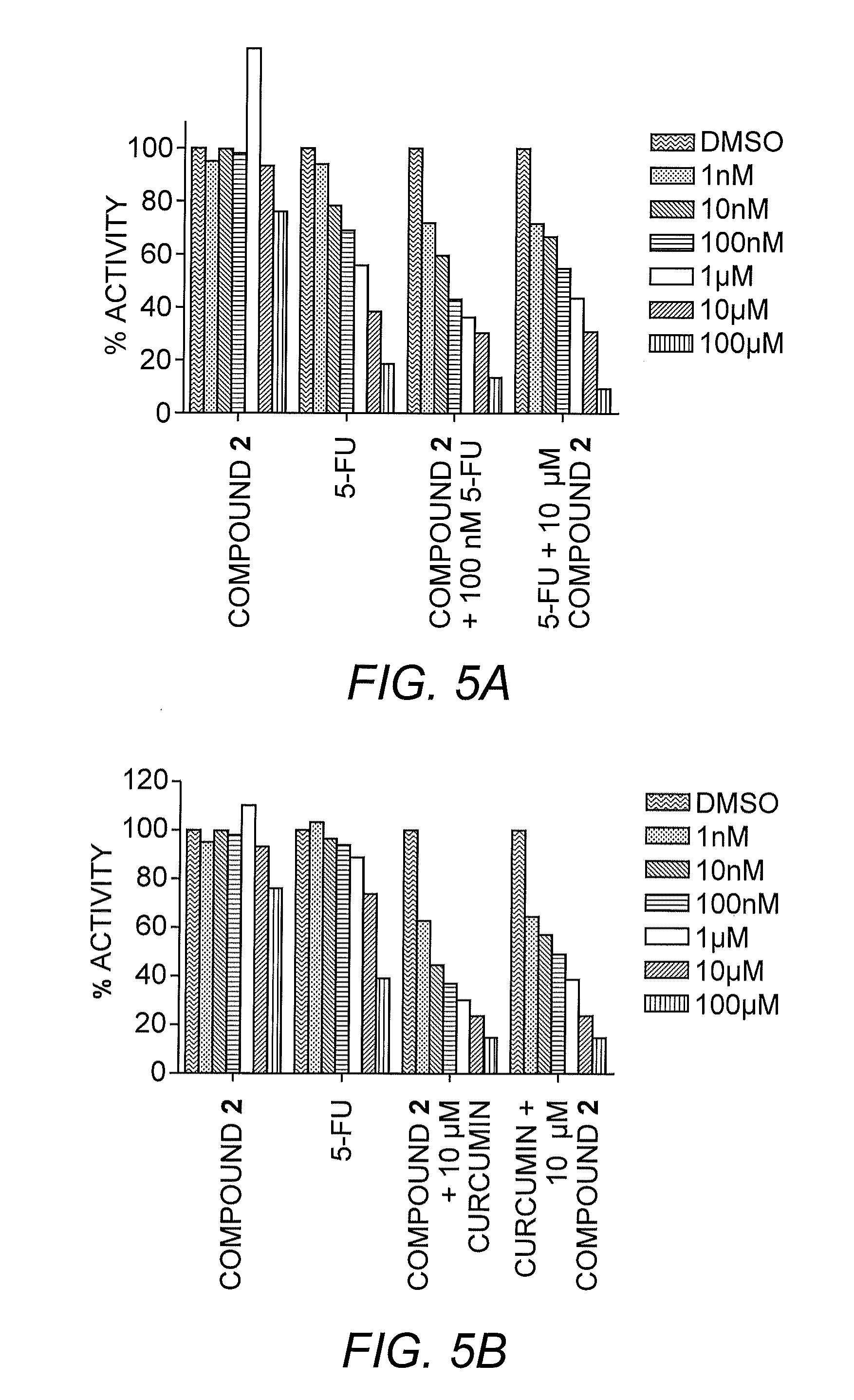
[0058]To illustrate the method of the present invention, a primary screen of >1000 compounds was conducted. The library was composed of a collection of novel compounds and the Library of Pharmacologically Active Compounds (LOPAC library, Sigma-Aldrich, St Louis, Mo.). All compounds were stored at a 10 mM concentration in DMSO to keep the DMSO concentration in the actual screen at 0.1% thereby minimizing toxicity. HeLa cells stably expressing the miR-reporter were treated with DMSO ranging from 0.1-1%. Luciferase signals were determined 48 hours after the treatment.
[0059]HeLa cells (2500 cells) were plated in each well of 384-well plate 24 hour before the addition of compounds. Compounds at 10 μM final concentration were added to each well. Luciferase signal were determined 48 hours after compound treatment. Using this screening assay, compound 1 was identified as a miR-21 antagonist.
[0060]This diazobenzene led to an increase of the luciferase sign...
example 3
Synthesis of the Diazobenzene 2
[0067]
[0068]4-Phenylazobenzoic acid (30 mg, 0.133 mmol) was dissolved in DCM (1 mL), followed by the addition of 1-ethyl-3-(3′-dimethylaminopropyl)carbodiimide (42 mg, 0.22 mmol) and hydroxybenzotriazole (21 mg, 0.15 mmol). Propargylamine (15 mg, 0.27 mmol) was added and the reaction was stirred at room temperature for 12 hours. The reaction was quenched with water (5 mL) and extracted with DCM (3×5 mL). The organic layer was dried with sodium sulfate, concentrated and purified by silica gel chromatography (2:1 hexane / ethyl acetate) to yield an orange solid (29 mg, 0.11 mmol, 86%). 1H NMR (400 MHz, CDCl3) δ 7.98-7.92 (m, 6H), 7.57-7.49 (m, 3H), 6.46 (bs, 1H), 4.29 (dd, J1=2.4 Hz, J2=4.8 Hz, 2H), 2.13 (t, J=2.4, 1H); 13C NMR (75 MHz, CDCl3) δ 166.6, 154.6, 152.7, 135.6, 131.9, 129.4, 128.3, 123.4, 123.2, 79.5, 72.4, 30.2.
[0069]HRMS Calcd for C16H14N3O (MH+): 264.1131, Found: 264.1135.
PUM
| Property | Measurement | Unit |
|---|---|---|
| green fluorescent | aaaaa | aaaaa |
| red fluorescent | aaaaa | aaaaa |
| yellow fluorescent | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com