Multiplex Cellular Assays Using Detectable Cell Barcodes
a multi-cellular assay and detection cell technology, applied in the field of multi-cellular assays using detectable cell barcodes, can solve the problems of insufficient system availability, inconvenient use, and difficulty in detecting cell barcodes, so as to reduce regent consumption, eliminate potential sample-to-sample variability, and improve the throughput of experiments
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
examples
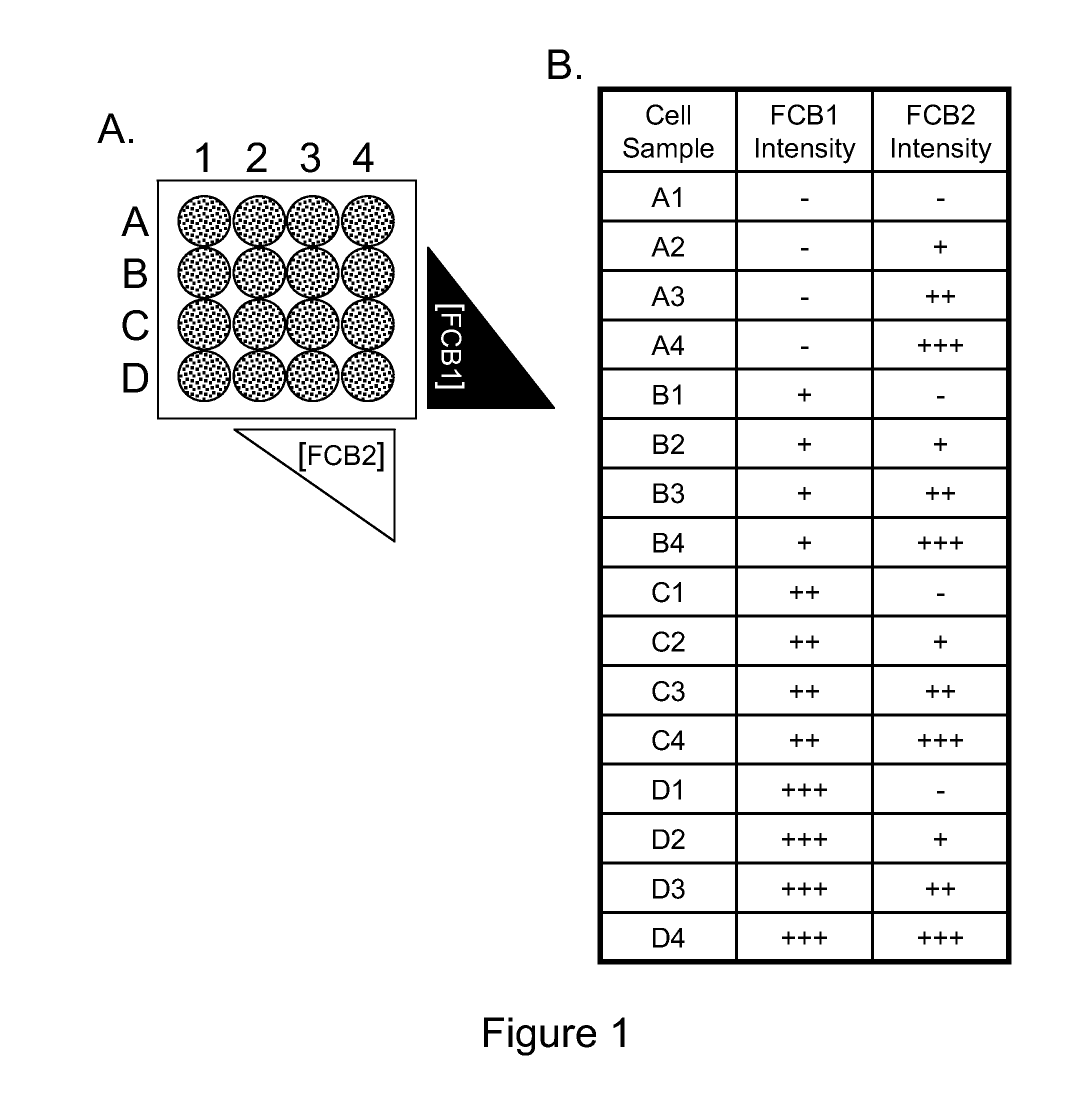
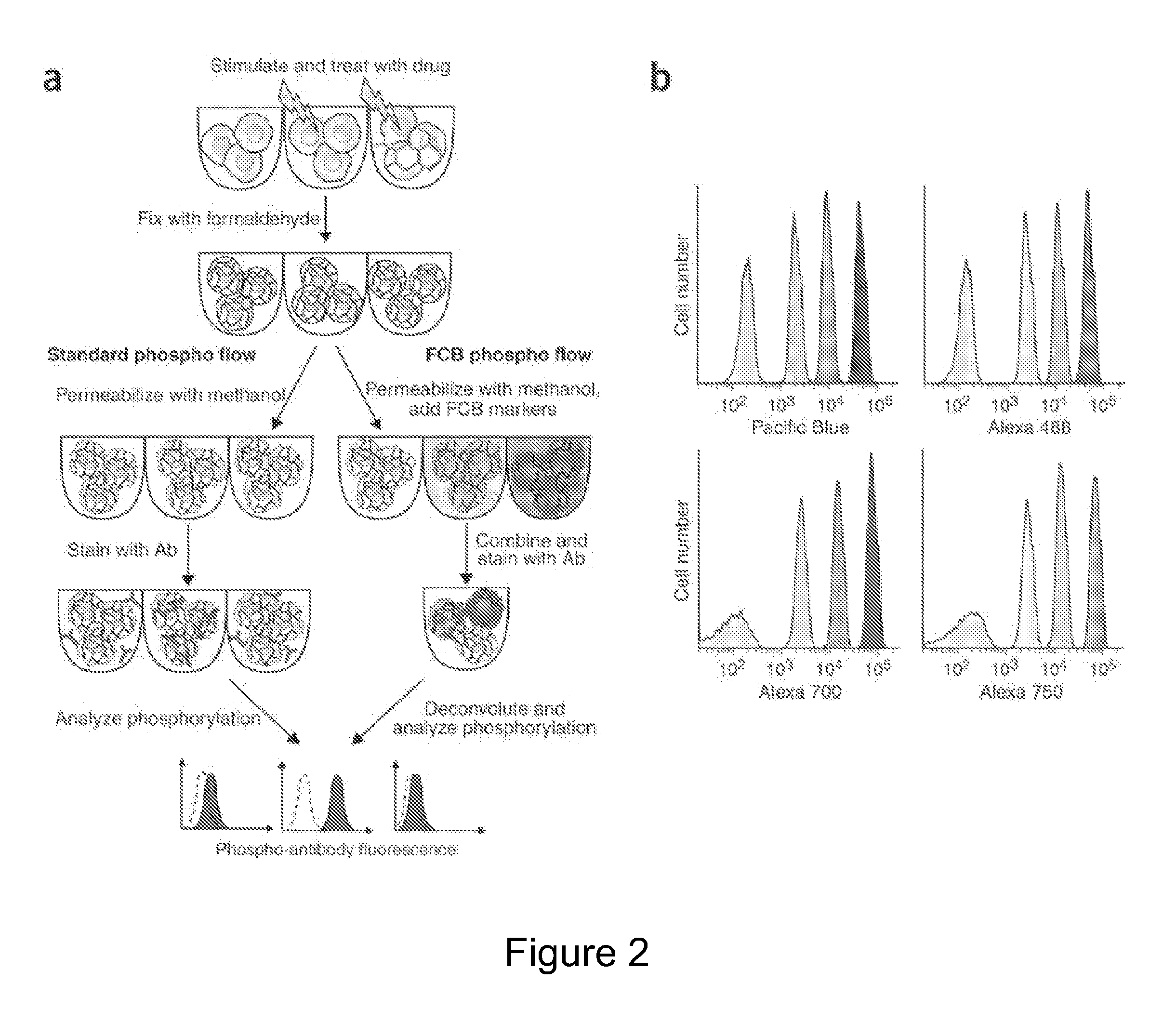
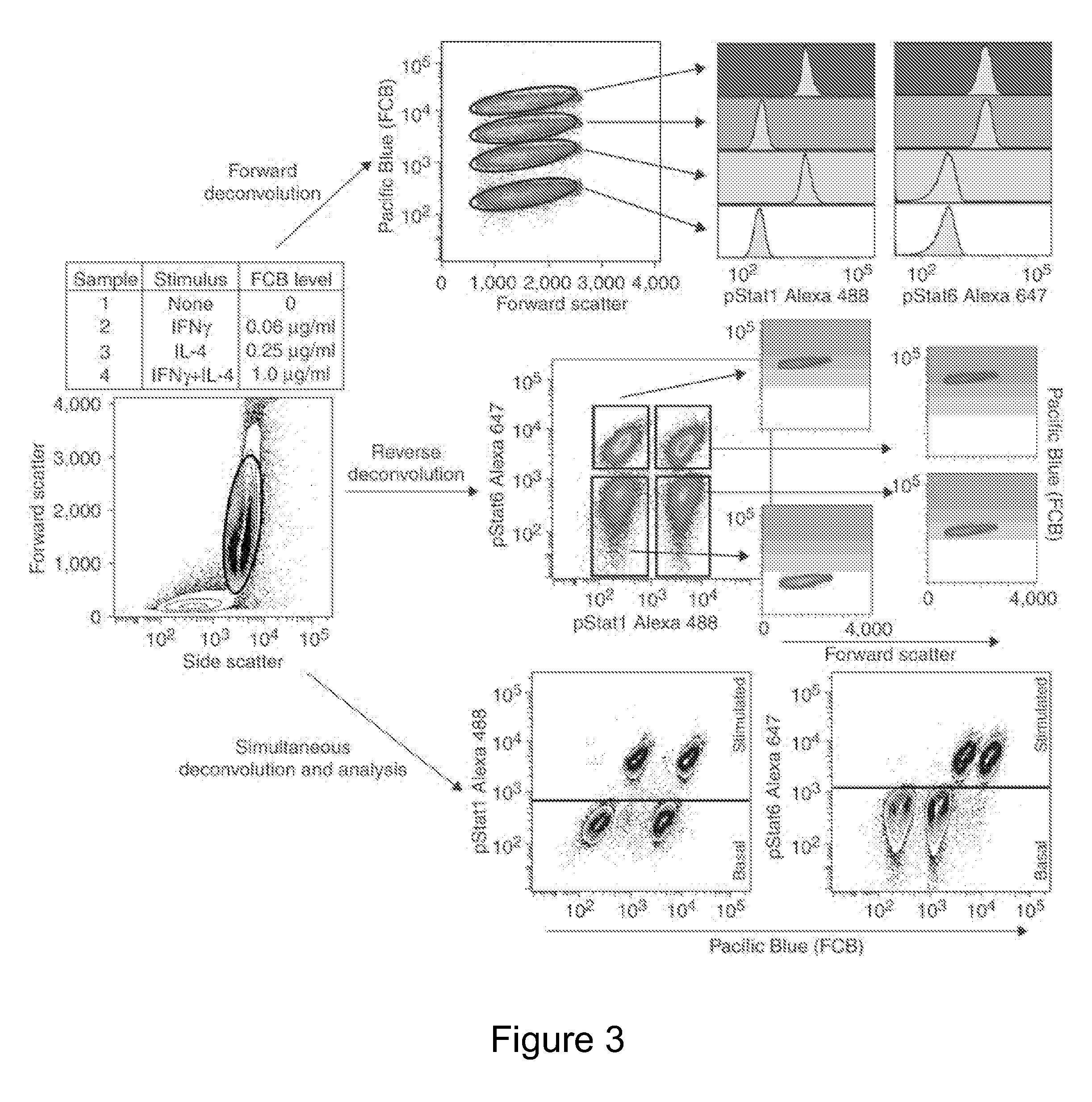
[0116]The following examples are put forth so as to provide those of ordinary skill in the art with a complete disclosure and description of how to make and use the subject invention, and are not intended to limit the scope of what is regarded as the invention. Efforts have been made to ensure accuracy with respect to the numbers used (e.g. amounts, temperature, concentrations, etc.) but some experimental errors and deviations should be allowed for. Unless otherwise indicated, parts are parts by weight, molecular weight is average molecular weight, and pressure is at or near atmospheric.
[0117]While the Examples below are described for FCBs, other DCBs may be employed in a similar manner. As noted above, the molecular mass-based DCB markers that are detected using mass spectrometry may also be employed in the multiplex cellular assays of the present invention. Exemplary methods for mass spectrometer analysis of cells can be found in the following: Ornatsky et al. “Multiple cellular a...
PUM
| Property | Measurement | Unit |
|---|---|---|
| concentrations | aaaaa | aaaaa |
| concentrations | aaaaa | aaaaa |
| concentrations | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com