Sequential sequencing
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
ization of the Human Oral Microbiome by Sequential Sequencing of Bacterial 16S Ribosomal DNA
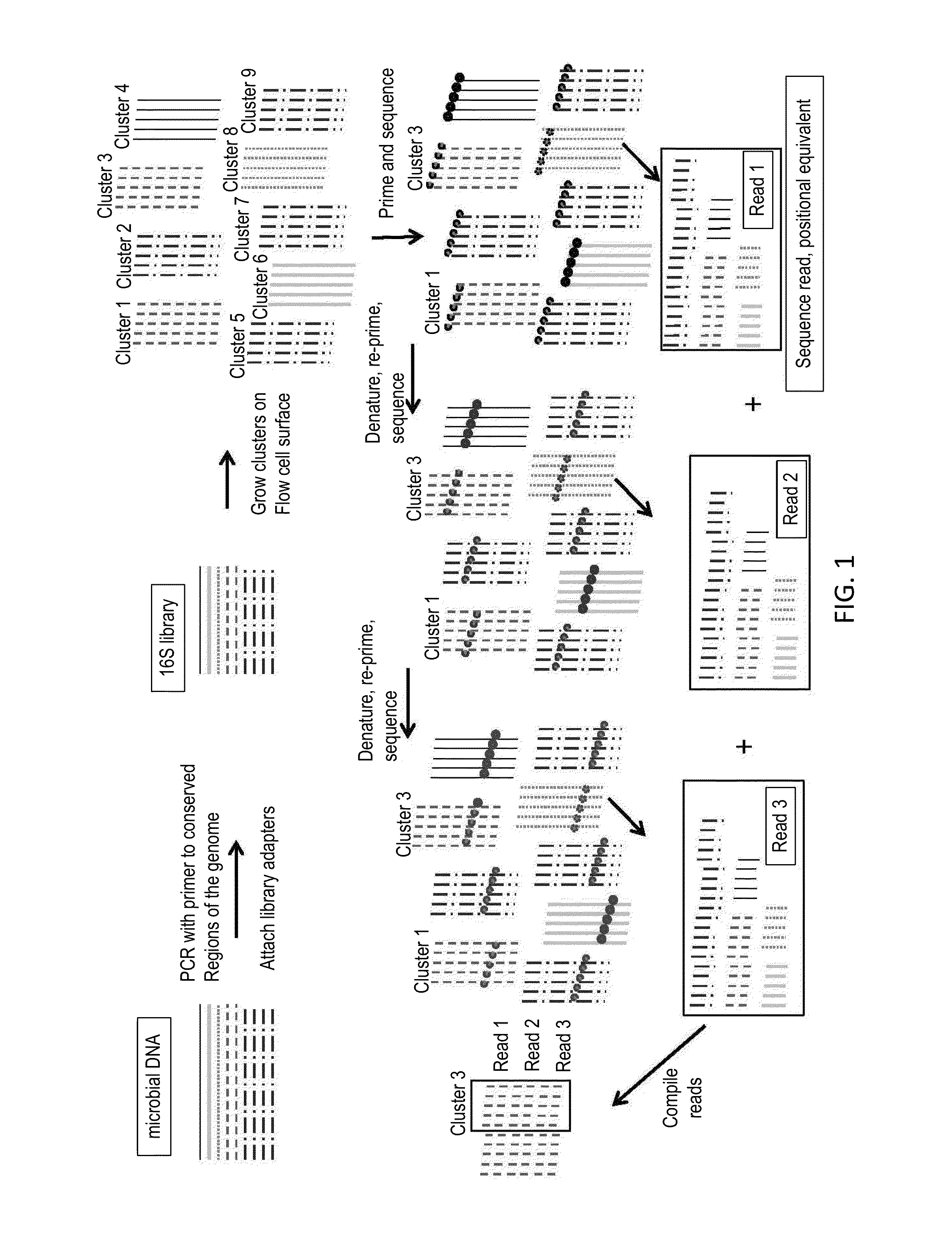
[0045]This example describes the characterization of the human oral microbiome by sequencing of the 16S rRNA gene sequences of a number of related bacterial organisms. 16S rRNA gene sequences contain species-specific hypervariable regions that can provide means for bacterial identification.
Sample nucleic acid
[0046]Microbial genomic DNA is isolated from human saliva using the OMNIgene-DISCOVER sample collection kit (DNA Genotek) according to the manufacturer's instructions. Extracted DNA is then fragmented via sonication to an average length of 400 by and purified using Agencourt AMPure XP beads (Beckman Coulter Genomics).
Generation of control and test 16 S libraries with ligated adapters
[0047]The NuGEN Ovation Ultralow Library System (NuGEN Technologies) is used to generate two directional next generation sequencing libraries from 100 ng of the purified sample according to manufacturer's inst...
example 2
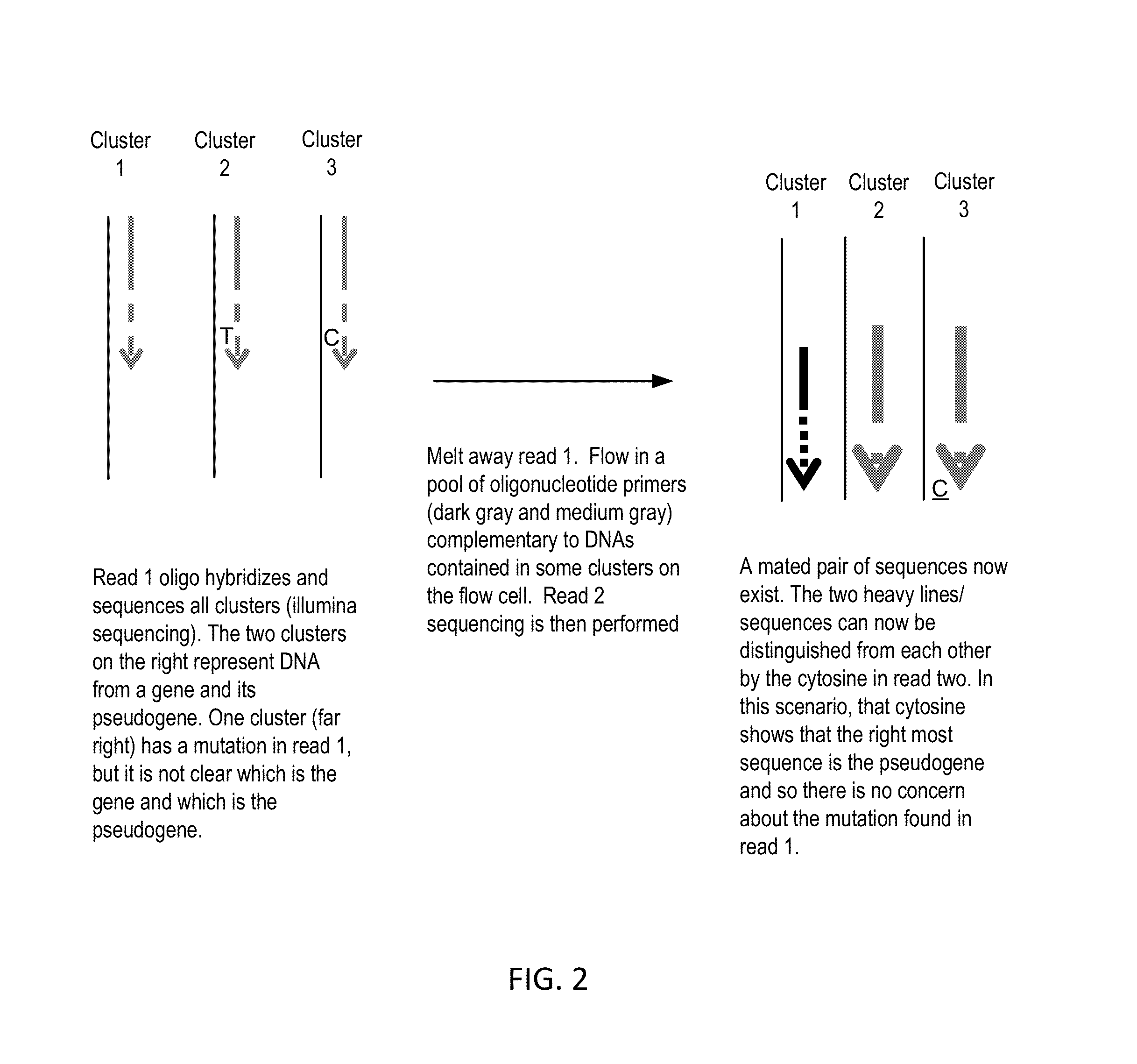
NA Sequencing—Distinguishing Between the SMN1 Gene and SMN2 Pseudogene Using Sequential Sequencing
[0050]Genomic DNA sequencing libraries are made using the NuGEN's Encore system. These libraries are sequenced on a DNA sequencing system such as those made by Illumina, Ion Torrent, Pacific Biosciences, or Complete Genomics. Following a first sequencing read, the DNA is denatured to wash away the first strand. A pool of primers that hybridize to common sequences in gene / pseudogene pairs are injected into the sequencer to act as a priming site for a second sequencing read. A primer set may include primers that will sequence through one of the nucleotide differences between SMN1 and SMN2 as well as primers that will generate sequence to read nucleotide differences, and therefore determine whether a sequencing read is from a globin gene or pseudogene. A combination of such primers will allow multiple gene / pseudogene pairs across the genome to be analyzed simultaneously for genetic mutatio...
example 3
[0051]A targeted DNA sequencing library is made using the a target enrichment product from NuGEN, Agilent, Illumina, or Nimblegen. These libraries are sequenced on a DNA sequencing system such as those made by Illumina, Ion Torrent, Pacific Biosciences, or Complete Genomics. Following a first sequencing read, the DNA is denatured to wash away the first strand. A pool of primers that hybridize to common sequences in gene / pseudogene pairs are injected into the sequencer to act as a priming site for a second sequencing read. A primer set may include primers that will sequence through one of the nucleotide differences between SMN1 and SMN2 as well as primers that will generate sequence to read nucleotide differences, and therefore determine whether a sequencing read is from a globin gene or pseudogene. A combination of such primers will allow multiple gene / pseudogene pairs across the genome to be analyzed simultaneously for genetic mutations. This type of technolog...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com