Bispecific antibodies and uses thereof
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
Expression of Anti-CD3 Fab Double Mutant in E. coli
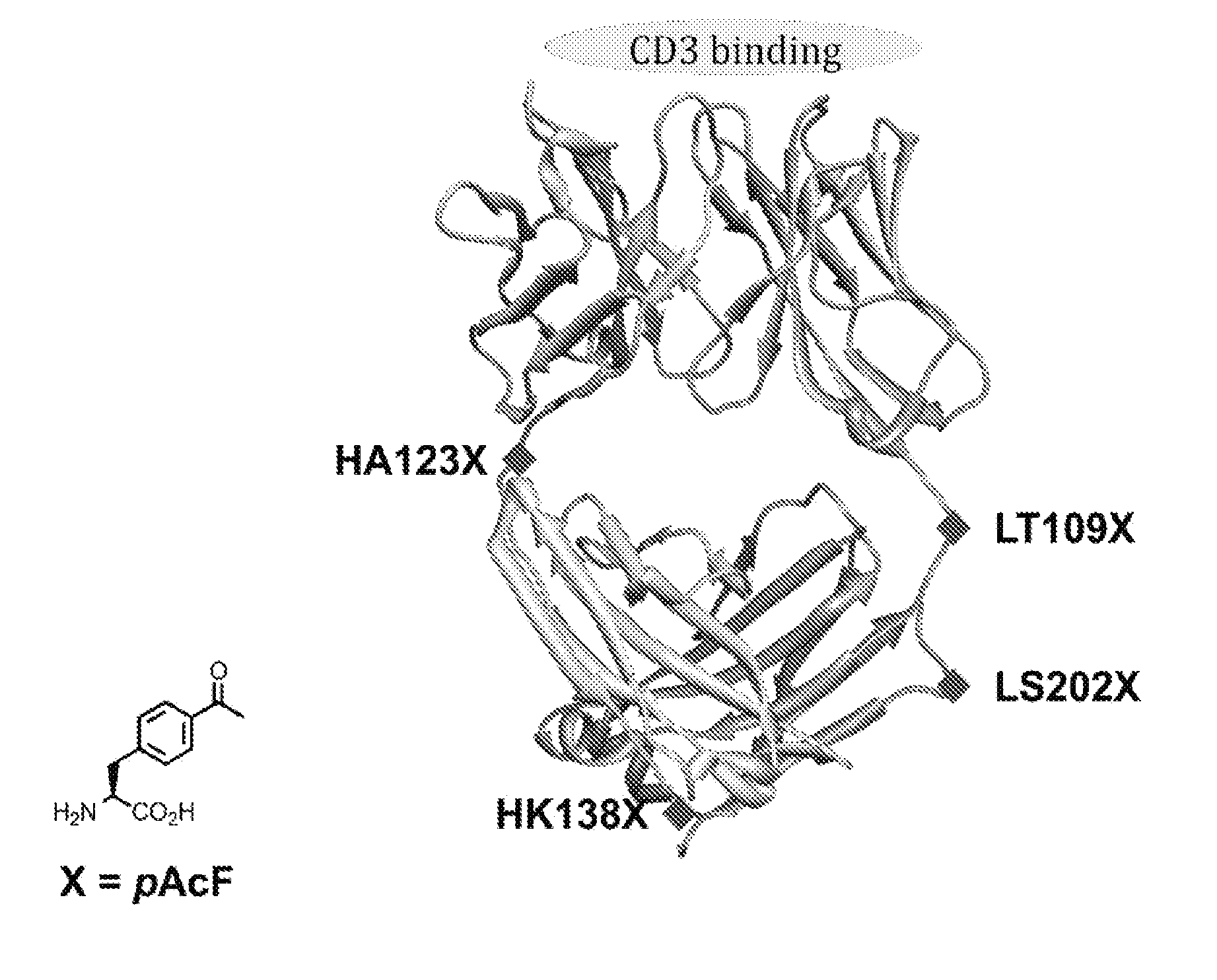
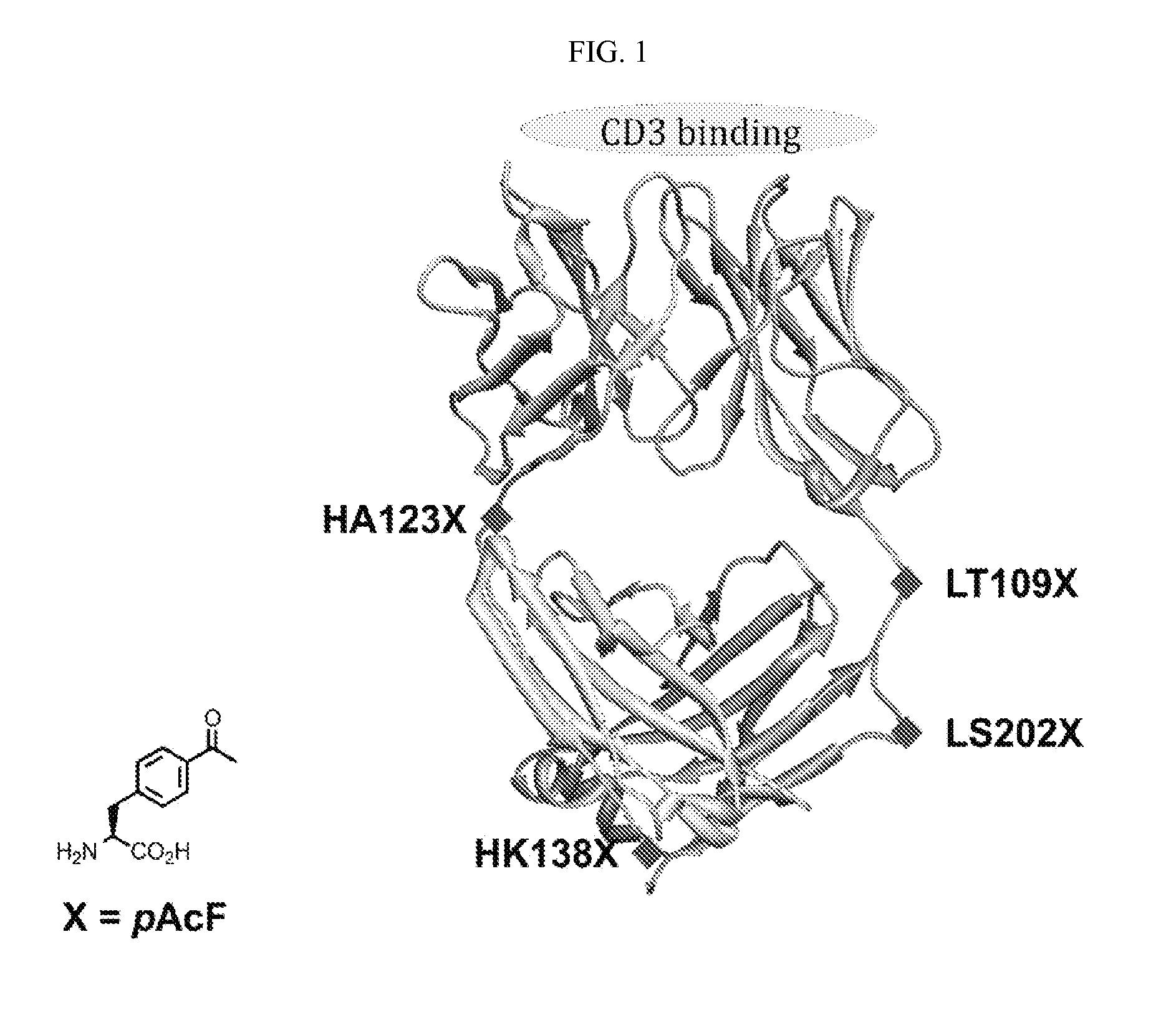
[0234]UCHT1-Fab sequences were obtained from the literature and inserted into a pBAD vector behind the stII signal sequence. The sites for unnatural amino acid incorporation (light chain threonine 109 (LC-Thr109), light chain serine 202 (LC-Ser202), heavy chain alanine 123 (HC-A123), and heavy chain lysine 138 (HC-Lys138)) were quickchanged (QuikChange® Stratagene) to TAG amber nonsense codon. For the double mutant, two residues (LC-Ser202 and HC-Lys138) were mutated to TAG amber nonsense codon. FIG. 1 depicts a ribbon diagram of the UCHT1-Fab fragment. The pBAD vector was co-transformed with pULTRA-pAcF (a vector containing orthogonal M. jannaschii tRNA and aminoacyl-tRNA synthetase specific for pAcF) in DH10B cells. The cells were grown in LB media (2 L) supplemented with 100 mg / ml ampicillin, 25 mg / ml chloramphenicol, and 1 mM pAcF at 37° C. and 250 rpm. At OD600 0.8, cells were induced with 0.2% arabinose and moved to 30° C. fo...
example 2
Synthesis of Bispecific Antibodies Using Genetically Encoded Unnatural Amino Acids
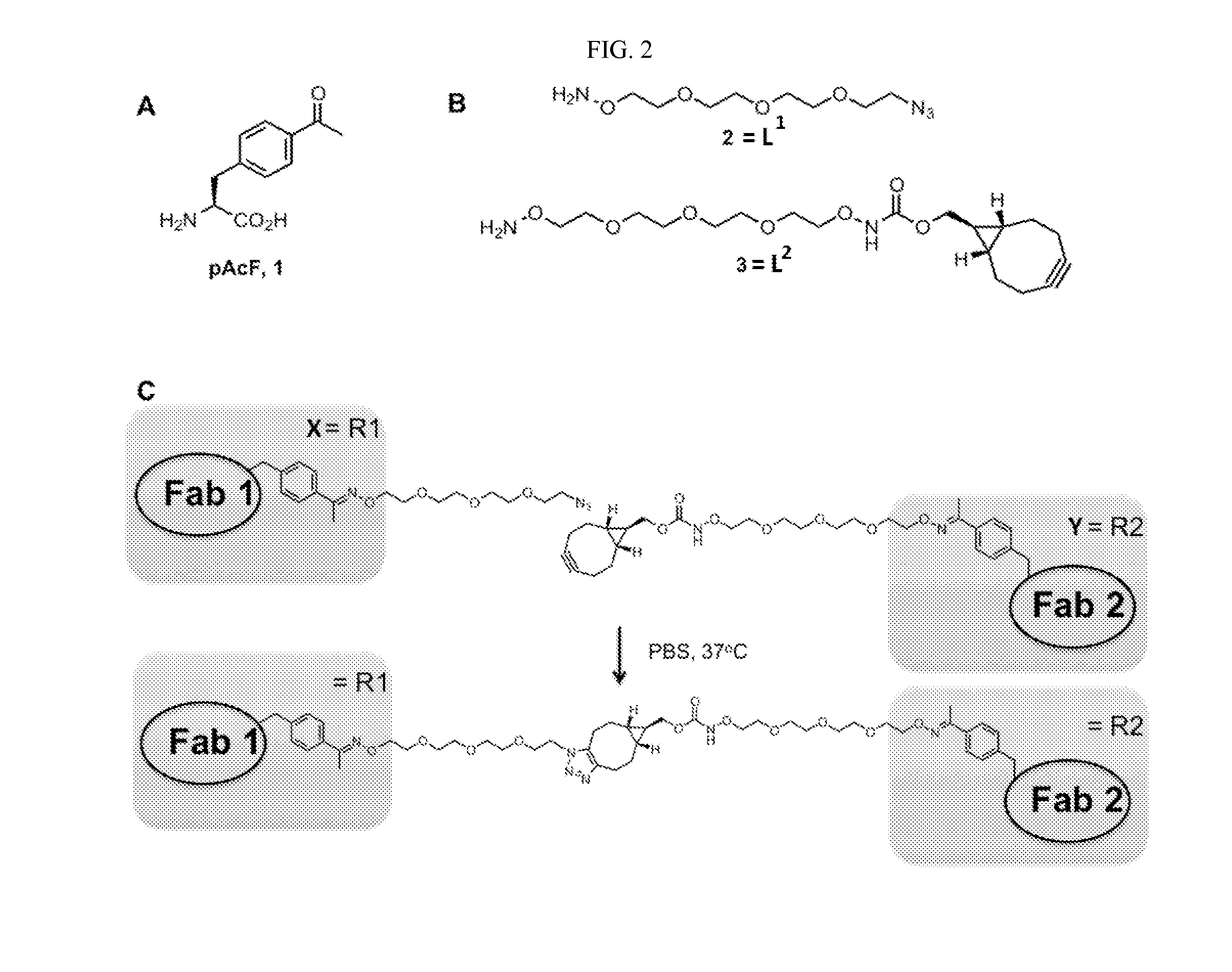
[0235]Our strategy takes advantage of genetically encoded unnatural amino acids with orthogonal chemical reactivity relative to the canonical twenty amino acids to site-specifically modify antibody fragments. Specifically, we used an evolved tRNA / aminoacyl-tRNA synthetase pair to site-specifically incorporate p-acetylphenylalanine (pAcF, FIG. 2A) at defined sites in each of two Fab fragments in response to an amber nonsense codon. The mutant Fab fragments were then selectively coupled by stable oxime formation using the alkoxy-amine termini of two bifunctional linkers (FIG. 2B). In a second step, the two Fab-linker conjugates were linked to each other in order to obtain the heterodimer through a copper-free [3+2] Huisgen-cycloaddition (“Click” reaction) (FIG. 2C). This approach has a number of advantages over recombinant technologies and conventional coupling chemistries. For example, the use of bioort...
example 3
Synthesis of Anti-Her2 / Anti-CD3 Heterodimer
[0238]pAcPhe was substituted at HC-K138 in the anti-anti-CD3 antibody, UCHT1. This site is distal to the antigen binding site and, when conjugated to the LC-5202 mutant anti-HER2 Fab with the same polyethylene glycol linker used above, should be long and flexible enough to allow the resulting bispecific antibody to productively bind both a CD3 positive T-cell and the HER2 positive target cell simultaneously. UCHT1 Fab was expressed in E. coli by the same method as described for anti-HER2 Fab and in Example 2, and the cyclooctyne linker-modified anti-CD3 was prepared as described in Example 7 in >95% yield as confirmed by ESI-MS (FIG. 3E, 3J). The anti-CD3-Fab-cyclooctyne conjugate was then coupled to anti-HER2-Fab-azide conjugate as described in Example 7 in 70% percent yield (determined by SDS-PAGE and chromatographic separation) and purified from unreacted Fab monomers by Superdex-200 size exclusion column. FIG. 4A shows the SDS-PAGE of t...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Distance | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com