Aptamer and detection method for C-reactive protein
a technology of c-reactive protein and aptamer, which is applied in the field of aptamer and a detection method relating to creactive protein, can solve the problems of large variation, difficult preservation, and antibodies used in the detection method, and achieve the effect of high sensitivity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
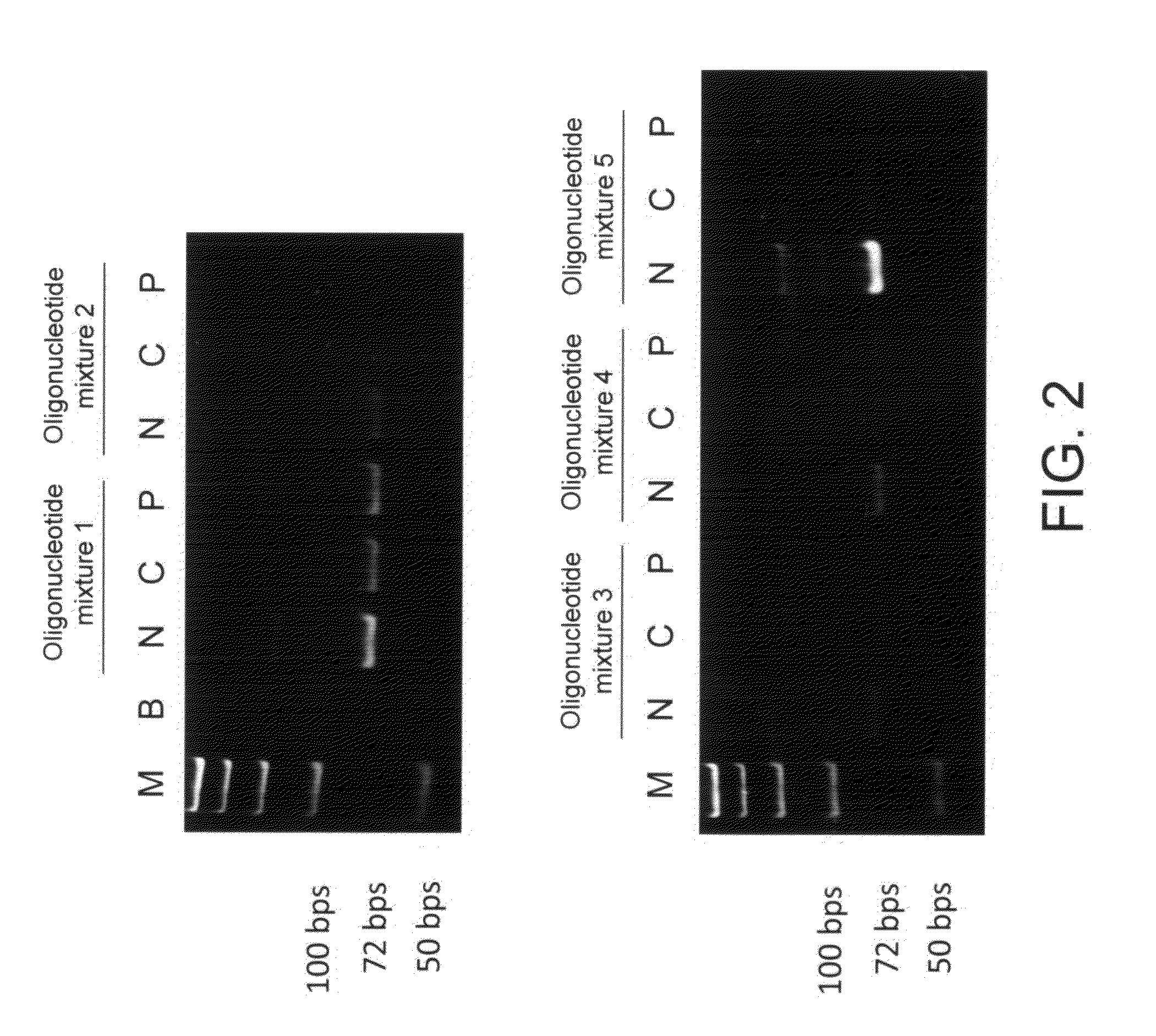
experiment 1
Initial Screening of Aptamers Having Affinity to CRPs
1. Establishment of Oligonucleotide Library
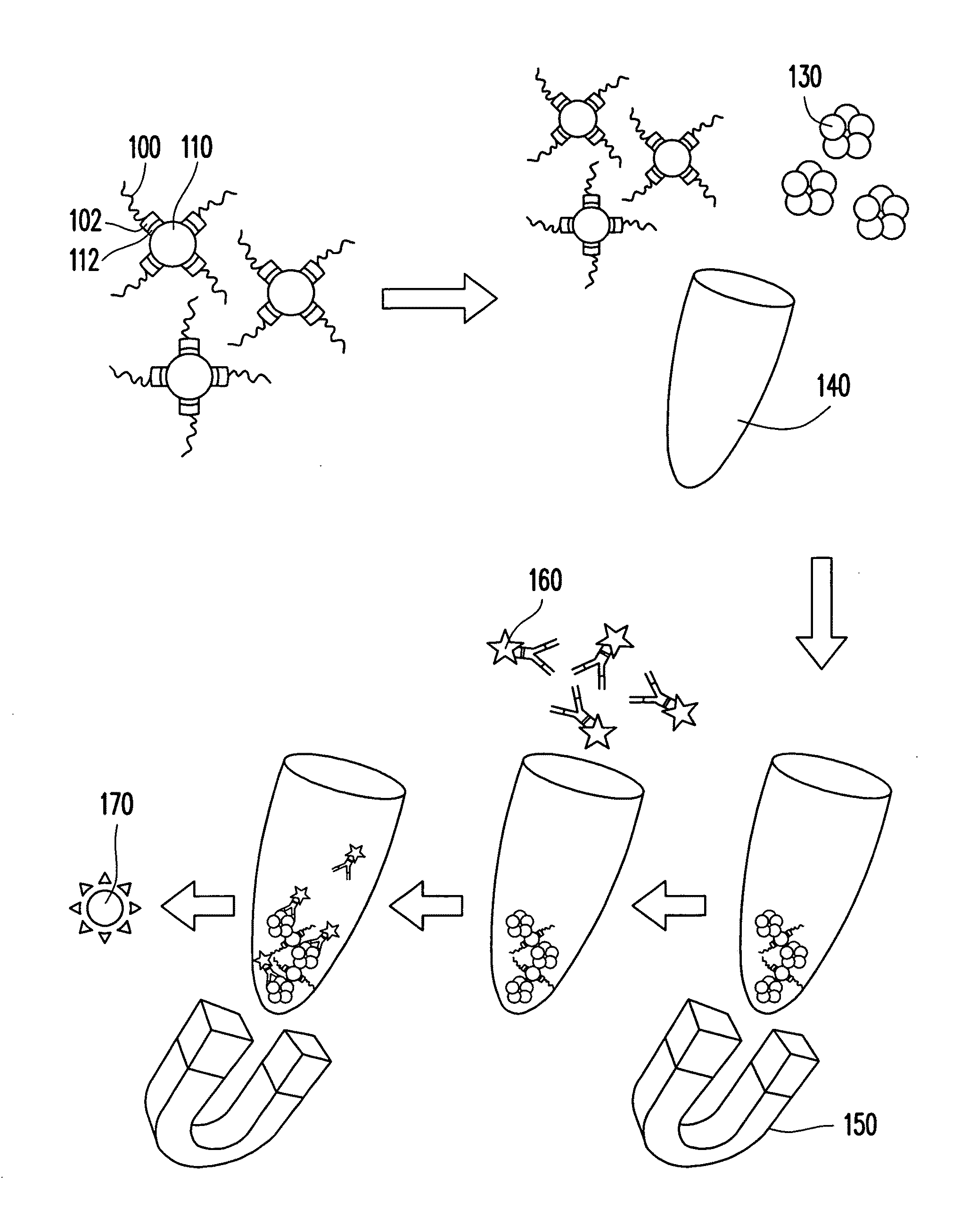
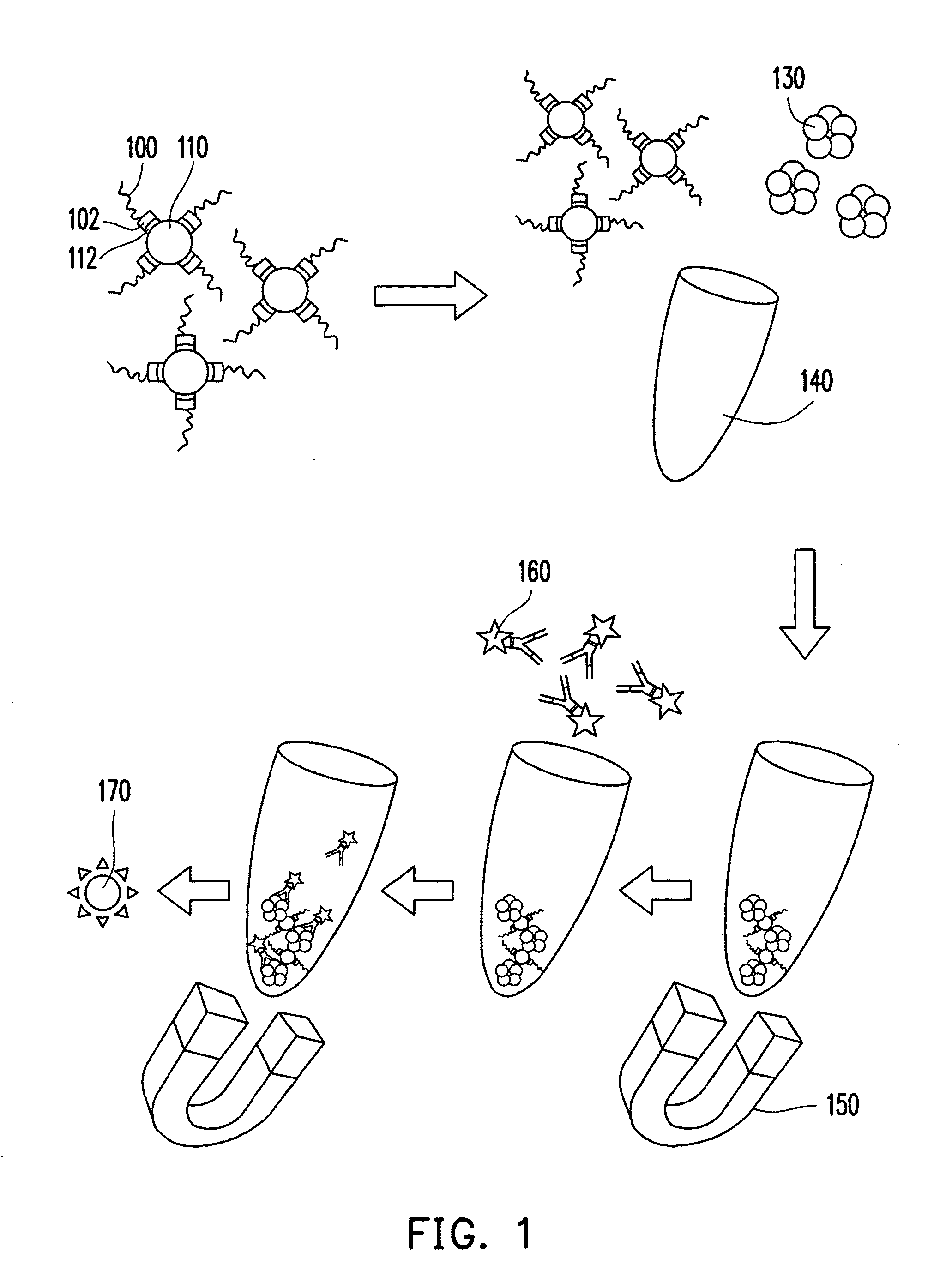
[0038]An oligonucleotide library includes 440 types of oligonucleotides. These oligonucleotides are synthesized by Medclub Scientific Co. Ltd., Taiwan, and each has a 72-mer nucleotide sequence shown in a SEQ ID NO:6. The 72-mer nucleotide sequence includes a random sequence constituted by 40 nucleotides (represented by n), a 5′-primer region constituted by 16 nucleotides, and a 3′-primer region constituted by 16 nucleotides:
[0039]5′-ggcaggaagacanaca-[n]40-tggtctgtggtgctgt-3′ (SEQ ID NO: 6). Here, n represents a nucleotide selected from adenine (a), thymine (t), cytosine (c), and guanine (g). The 5′-primer region and the 3′-primer region are respectively designed to be nucleotide sequences recognized by Super-Therm Gold DNA polymerase (Bertec Enterprise Co. Ltd., Taiwan) for performing a polymerase chain reaction (PCR).
[0040]Then, a suitable amount of oligonucleotide library is dissolved ...
experiment 2
Cloning of Aptamers Having Affinity to CRPs
[0049]The PCR product (1 μL) obtained from Experiment 1, pGEM®-T Easy vector (1 μL), 2× rapid ligation buffer (5 μL), and T4 DNA ligase (1 μL) are mixed well, and deionized water is added to make a total volume of the reaction to be 10 μL. The reaction is then placed on ice overnight. Afterwards, 50 μL of JM 109 Escherichia coli (E. coli) competent cells is added to the above solution and mixed well. The reaction is then placed on ice for 20 min. Next, the reaction undergoes heat shock for 45-50 sec in a 42° C. water bath and is rapidly transferred on ice for 2 min. Thereafter, 950 μL of SOC medium is added and incubated under 37° C. in a shaker for 1.5 hr at 150 revolution per min (rpm). The incubated product is well spread on an LB plate containing 1 mg / ML ampicillin, 0.5 mM isopropyl-β-D-thiogalactopyranoside (IPTG), and 80 μg / mL X-Gal, and incubated overnight at 37° C. Next, fifteen ampicillin-resistant colonies are picked from the LB p...
experiment 3
Screening of Aptamers Having Affinity to CRPs
[0050]Firstly, a 50 mg / L CRP solution and a 1% BSA solution are prepared respectively by dissolving human serum CRP and BSA in 0.05 M carbonate buffer. Next, 50 μL of the CRP solution obtained is added to a 0.2 mL PCR eppendorf and placed under 4° C. for coating overnight. The eppendorf with an inside wall coated with the CRPs is removed from 4° C. and washed with 0.01 M PBS for three times. Thereafter, 50 μL of 1% BSA solution is added and reacted under room temperature for 1 hr. Afterwards, the inside wall of the eppendorf is washed three times with 0.01 M PBS, and an eppendorf A with the inside wall coated with the CRPs is obtained for use.
[0051]In addition, 50 μL of the BSA solution obtained is added to a 0.2 mL PCR eppendorf and placed under 4° C. for coating overnight. The eppendorf with an inside wall coated with BSA is washed three times with 0.01 M phosphate buffer, and an eppendorf B with the inside wall coated with the BSA is o...
PUM
| Property | Measurement | Unit |
|---|---|---|
| concentration | aaaaa | aaaaa |
| concentration | aaaaa | aaaaa |
| concentration | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com