Penicillin binding protein from Streptococcus pneumoniae
a technology of penicillin and pneumoniae, which is applied in the field of recombinant dna technology, can solve the problems of mdr organisms that are a real threat to humans, children and the elderly, and emergence and rapid spread of beta-lactam resistance in streptococcus pneumoniae, and achieves the effects of reducing the number of mdr organisms, and reducing the number of mdl
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
example 1
Construction of a DNA Vector for Expressing Streptococcus pnuemoniae pbp-nv.sup.S Gene in a Heterolocous Host
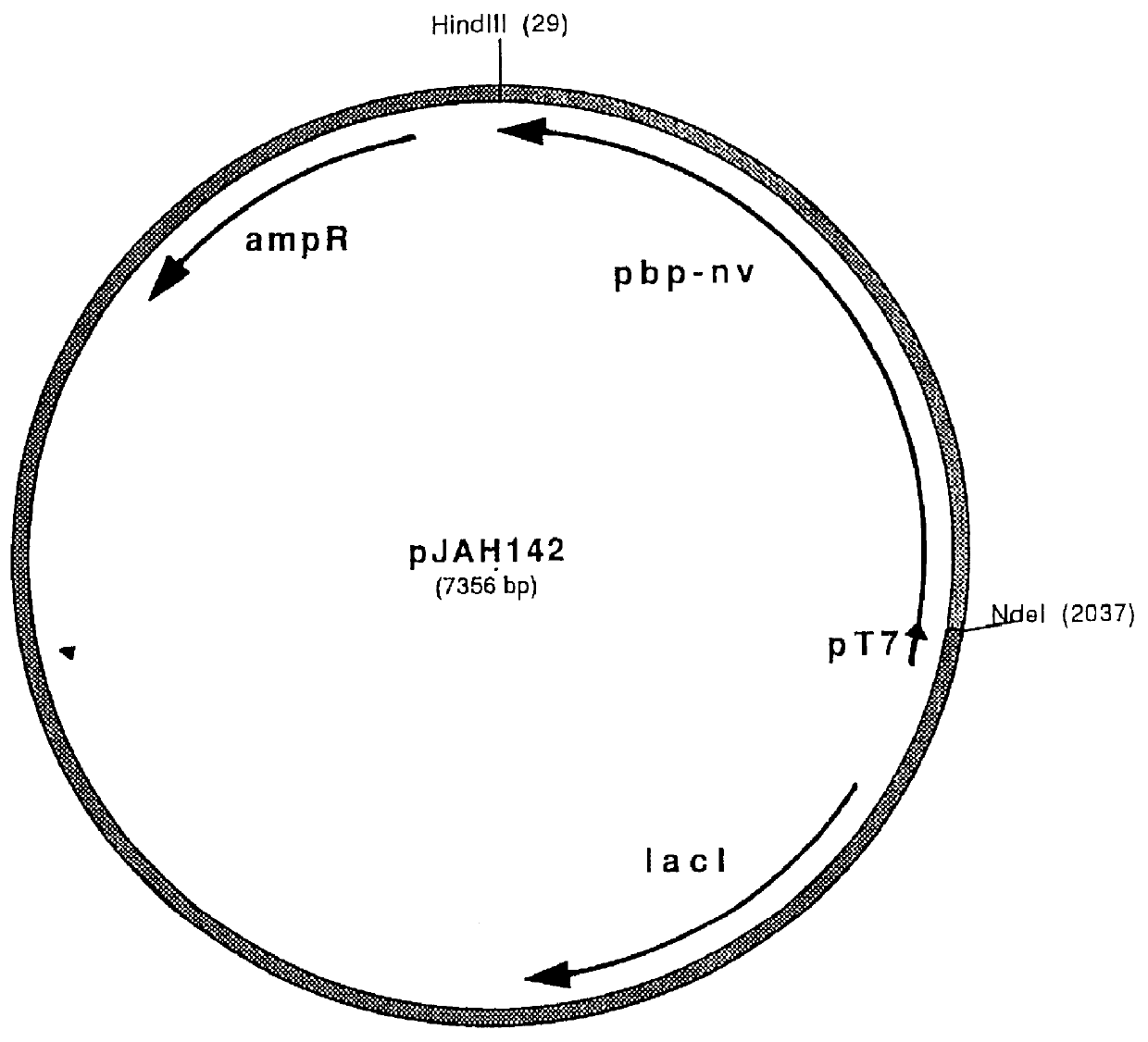
Plasmid JAH142 (See Figure) is an approximately 7300 base pair expression vector suitable for expressing a modified pbp-nv.sup.S in procaryotic host E. coli. This plasmid contains an origin of replication (Ori), an ampicillin resistance gene (Amp), useful for selecting cells that have incorporated the vector following a tranformation procedure, and further comprises the T7 promoter in operable linkage to the coding region of said PBP gene. Parent plasmid pET11A (obtained from Novogen, Madison, Wis.) was linearized by digestion with endonucleases NdeI and HindIII. Linearized pET11A was ligated to a DNA fragment bearing NdeI and HindIII sticky ends, comprising a modified pbp-nv.sup.s gene. The pbp-nv.sup.s gene ligated into pJAH142 was modified at the 5' end in order to simplify purification of the encoded protein product. For this purpose, an oligonucleotide encoding 8 histidi...
example 2
Construction of a DNA Vector for Expression of pbp-nv in a heteroloaous host
The plasmid construction method outlined in Example 1 is followed to construct a vector for expressing PBP-Nv in a heterologous host such as E. coli. A "his-tag" oligonucleotide comprising the same structure and sequence disclosed in Example 1 is inserted at the 3' end of the ATG initiation codon at nucleotide position 3 of SEQ ID NO.1.
example 3
Expression of Streptococcus pneumoniae pbp-nvS Gene in Echerichia coli
Expression plasmid JAH142 was transformed into E. coli BL21 (DE3) (F.sup.- ompT[lon]hsdS r.sub.B.sup.- m.sub.B.sup.-) using standard methods (See e.g. Sambrook et al. Supra). Transformants, chosen at random were tested for the presence of pJAH142 by agarose gel electrophoresis using quick plasmid preparations. Id. Transformants were grown overnight at 37.degree. C. in LB medium supplemented with 100 .mu.g / ml ampicillin. The overnight culture was diluted into fresh LB medium and allowed to grow to an O.D..sub.600 of 0.4 to 0.6. At that point, expression of the vector-bound pbp-nvS gene was induced by adding 0.4 mM IPTG for a period of 3 hours. The induced-culture was then pelleted by centrifugation in preparation for protein purification (See Example 4).
PUM
| Property | Measurement | Unit |
|---|---|---|
| pH | aaaaa | aaaaa |
| bed-volume | aaaaa | aaaaa |
| pH | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com