Pig carcass character GFAT1 gene clone and its application
A pig carcass and gene technology, applied in the fields of application, genetic engineering, plant genetic improvement, etc., can solve the problems of limited, limited number of new genes, few genes, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
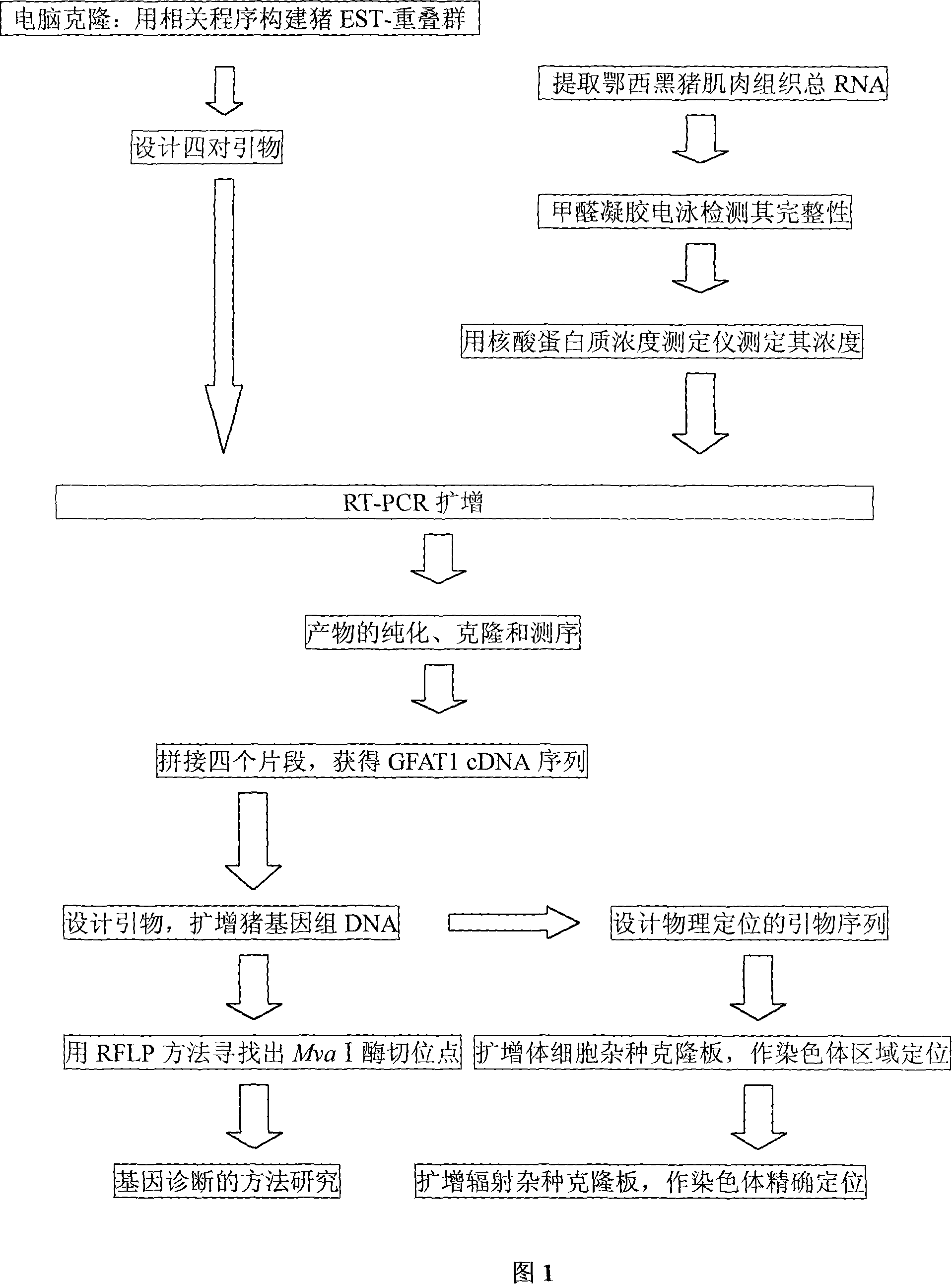
[0047] 1. Cloning of GFAT1 gene:
[0048] (1) Primer design:
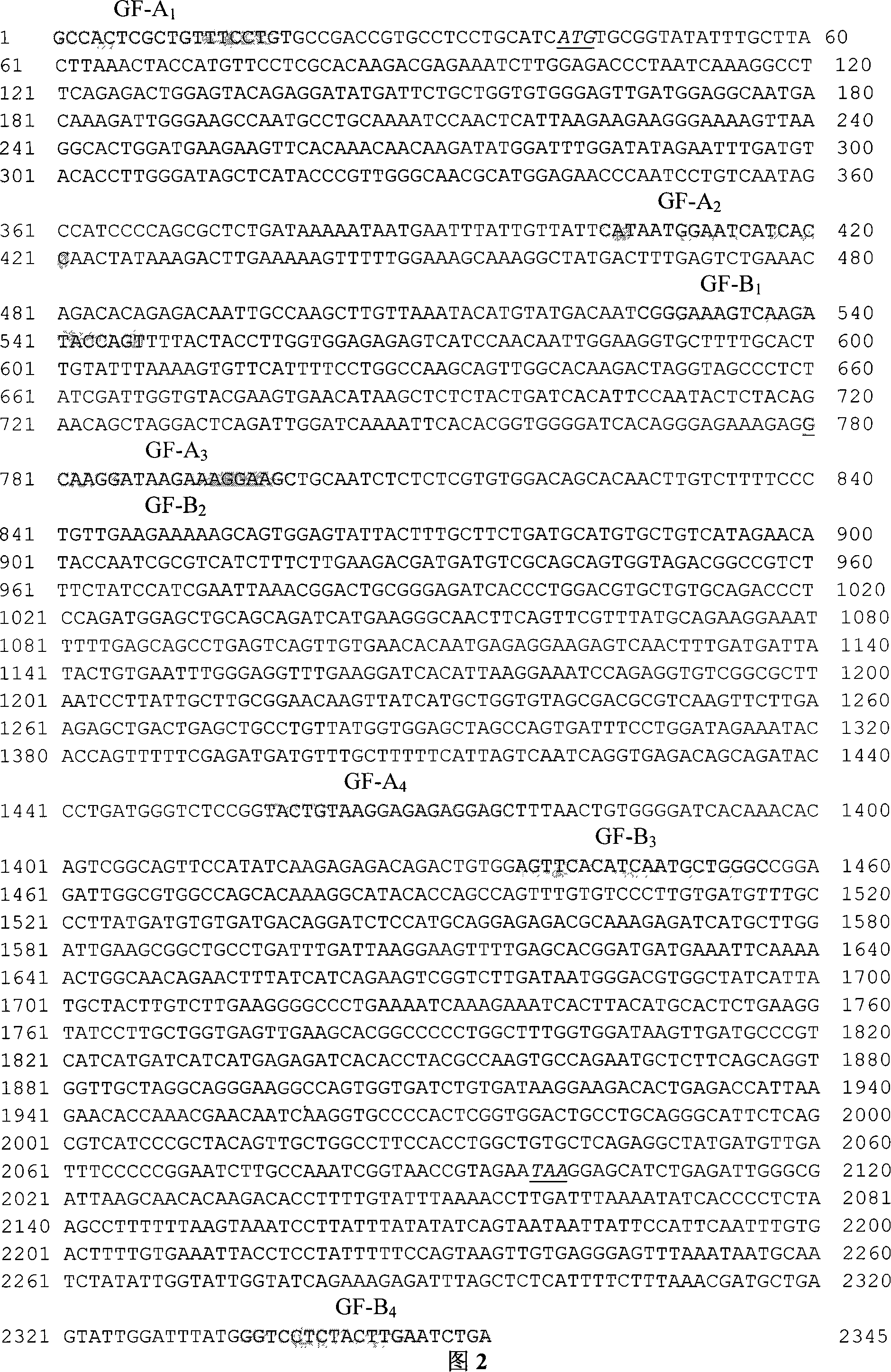
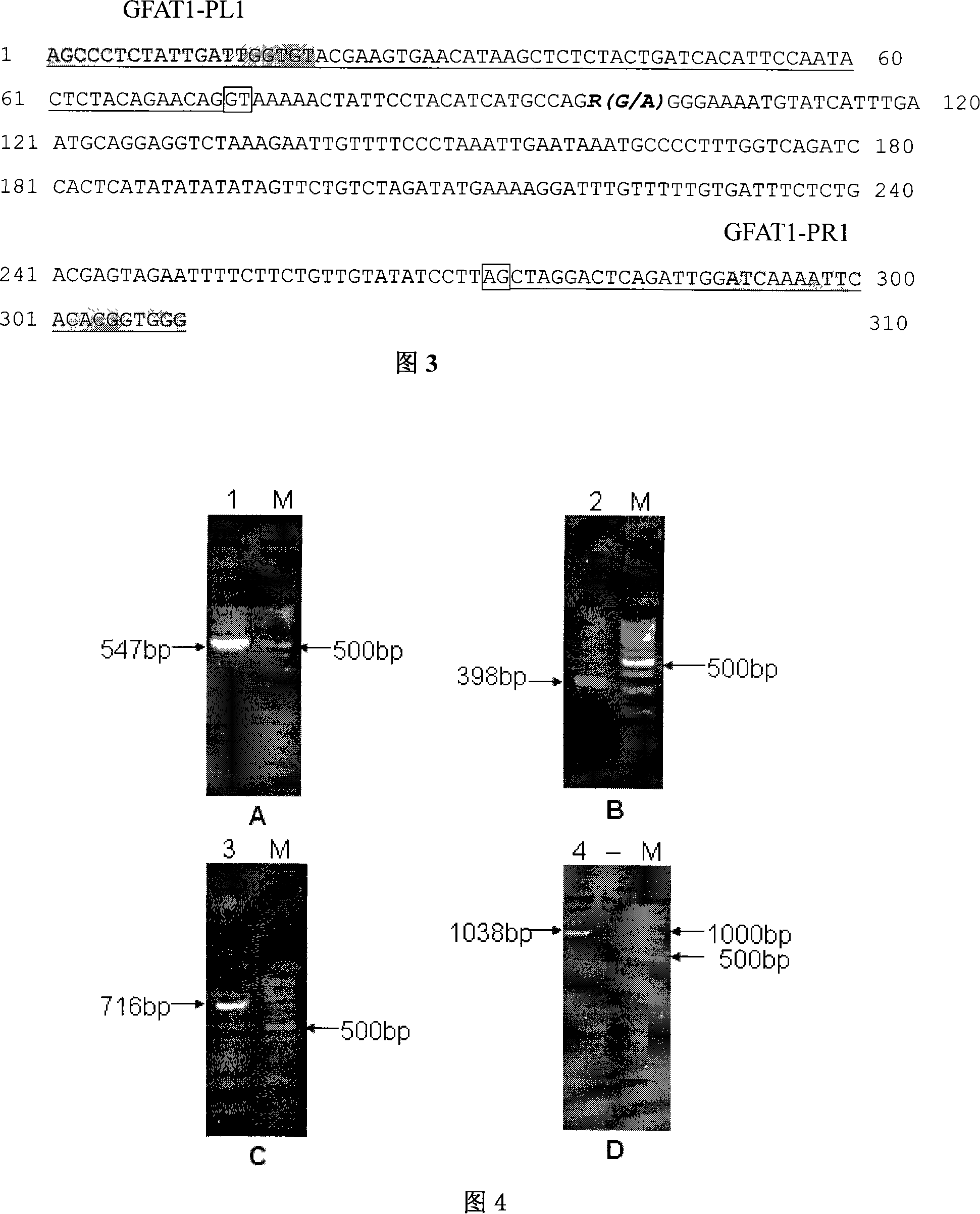
[0049] Using human GFAT1 gene cDNA (GenBank accession number: NM_002056) as an information probe, using the BLAST tool in NCBI to perform homologous sequence screening in the GenBank pig EST database, and obtain a series of ESTs with a homology of more than 85% (the fragment length is greater than 100bp), query the corresponding sequences with the accession numbers of these ESTs in NCBI with ENTREZ (http: / / www.ncbi.nlm.nih.gov / Web / Search / index.html), and then use the ASSEMBLY program in GeneTool to construct pig EST-contig. Design four pairs of primers GF-A according to EST splicing sequence 1 and GF-B 1 , GF-A 2 and GF-B 2 , GF-A 3 and GF-B 3 , and GF-A 4 and GF-B 4 Amplified to obtain four fragments A 1 B 1 ;A 2 B 2 ;A 3 B 3 and A 4 B 4 (as shown in Table 3). The PCR product was recovered and purified, cloned and sequenced, and the sequence was spliced to obtain the complete coding region sequ...
Embodiment 2
[0098] The distribution situation of embodiment 2 PCR-RFLP-MvaI polymorphism in each pig breed
[0099] Five pig herds were detected by PCR-MvaI-RFLP: Tongcheng purebred (Chinese local pig blood) population, large white purebred (foreign blood), Landrace purebred (foreign pig blood), Changtong (Chinese and foreign hybrid pig blood) And Dachangtong (Chinese-foreign hybrid pig bloodline), the gene frequency of each pig group is shown in Table 6, and the genotypes of GFAT1 gene are distributed in Landrace and Dachangtong pigs, and GG was not detected in Large White and Changtong pigs Homozygous genotype, only AA homozygous genotype in Tongcheng pigs.
[0100] Table 6 GFAT1 gene PCR product genotype frequency and gene frequency
[0101]
Embodiment 3
[0102] Example 3 Application of Association Analysis of Pig Marker Traits
[0103] The association analysis between MvaI-RFLP polymorphism in the 8th intron of pig GFAT1 gene and some production traits was carried out in a Tongcheng pig herd. The analysis results showed that the carcass length and fat thickness at the midpoint of the gluteus medius were significantly different between GA and GG genotypes (P<0.05) (Table 7)
[0104] It can be seen from Table 7 that the G allele is a favorable marker for carcass straight length and fat thickness at the midpoint of the gluteus medius (long carcass, thin rump fat), and GG genotype markers can be used to increase body length and reduce gluteus medius at the same time. The selection mark for the fat thickness at the point.
[0105] Table 7 Association analysis of different GFAT1 gene MvaI-RFLP genotypes and some production traits
[0106] genotype
[0107] Note: The same letter in the same column means no significant dif...
PUM
| Property | Measurement | Unit |
|---|---|---|
| molecular weight | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com