Human nucleotide repair protein gene main polymorphism site fast detection method and kit thereof
A polymorphic site and protein repair technology, applied in the field of molecular biology, can solve the problems of time-consuming and cumbersome operation, poor specificity and sensitivity, time-consuming and expensive sequencing and chip technology, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
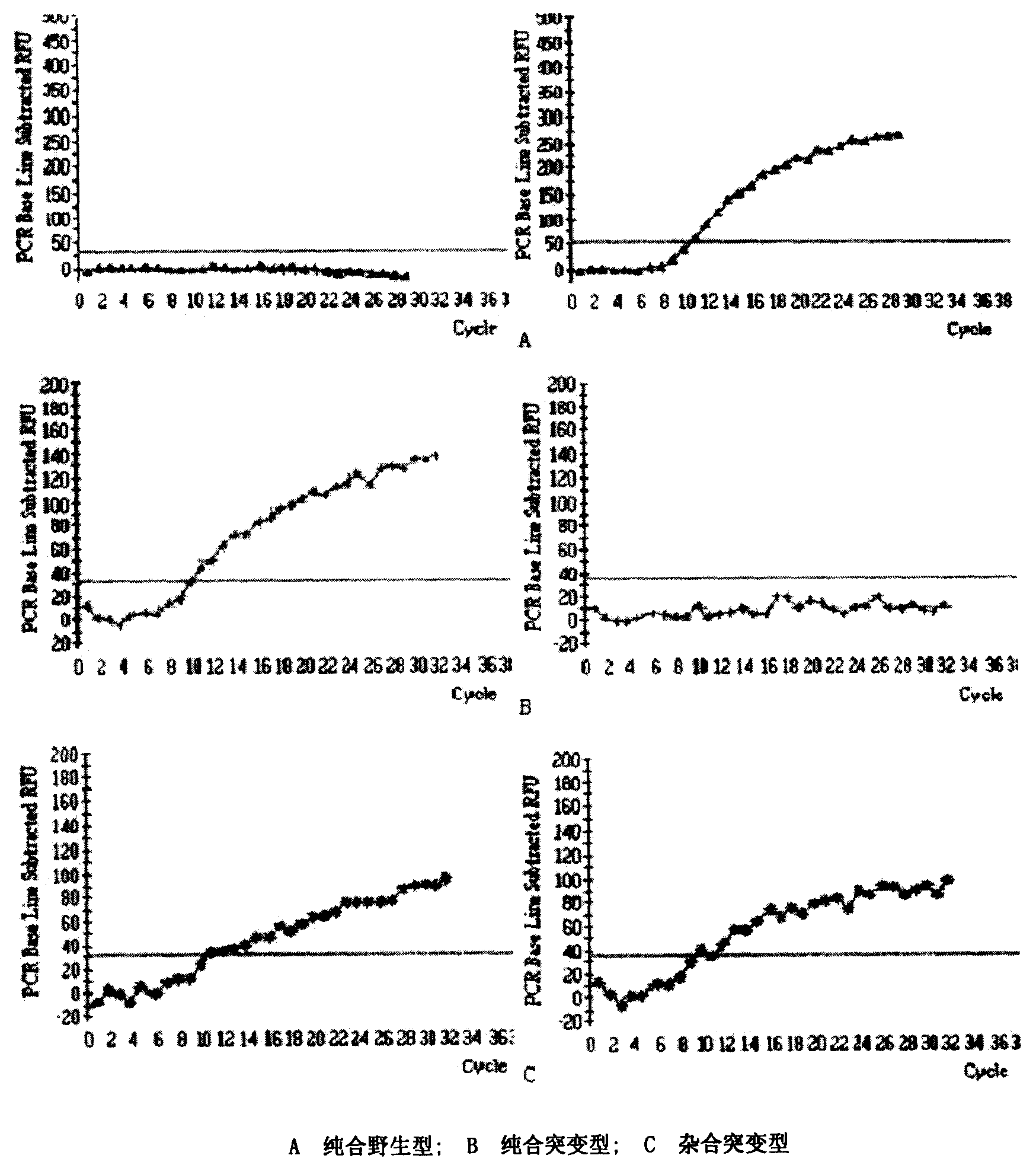
Image
Examples
Embodiment Construction
[0033] The composition of the kit of the present invention is as follows:
[0034] Reaction mixture: 1×PCR buffer (0.1% TritonX-100; 2.0~6.0mmol / L MgCl 2 ; 8mmol / L(NH 4 ) 2 SO 4 ; 50mmol / L KCl; 10mmol / LTris-HCl(pH8.0)); contains two primers each 0.3μmol / L, Probe-wt0.2μmol / L, Probe-mu0.3μmol / L; dNTP each 250μmol / L, HotTaq enzyme 1.0U, total reaction volume 50μl.
[0035] Sample extraction solution: (1). Absolute ethanol; (2). 1.5% triethanolamine lauric acid sulfate (TLS); (3). Chloroform: isoamyl alcohol (24:1); (4). TE (pH 8 .0).
[0036] R194W mutant wild-type probe sequence:
[0037] Probe-wt: FAM-5’-GCGCTCAGCCGGATCAACAAGACAGCGC-3’-DABCYL
[0038] R194W mutant mutant probe sequence:
[0039] Probe-mu: HEX-5’-GCACGGCTCTCTTCTTCAGCTGGATCGTGC-3’-DABCYL
[0040] R280H mutant wild-type probe sequence:
[0041] Probe-wt: FAM-5’-CGCTGCCAGCTCCAACTCGTACCAGCG-3’-DABCYL
[0042] R280H mutant mutant probe sequence:
[0043] Probe-mu: HEX-5’-CGCTGCCAGCTCCAACTCATACCAGCG-3’-DABCYL
[0044] R399Q...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com