Rice HAP3 and application of the same in improving stress tolerance of plants
A gene and plant technology, applied in the application field of rice HAP3 and its gene in improving plant stress tolerance, to achieve the effects of improving stress tolerance, improving tolerance, and enhancing salt resistance
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
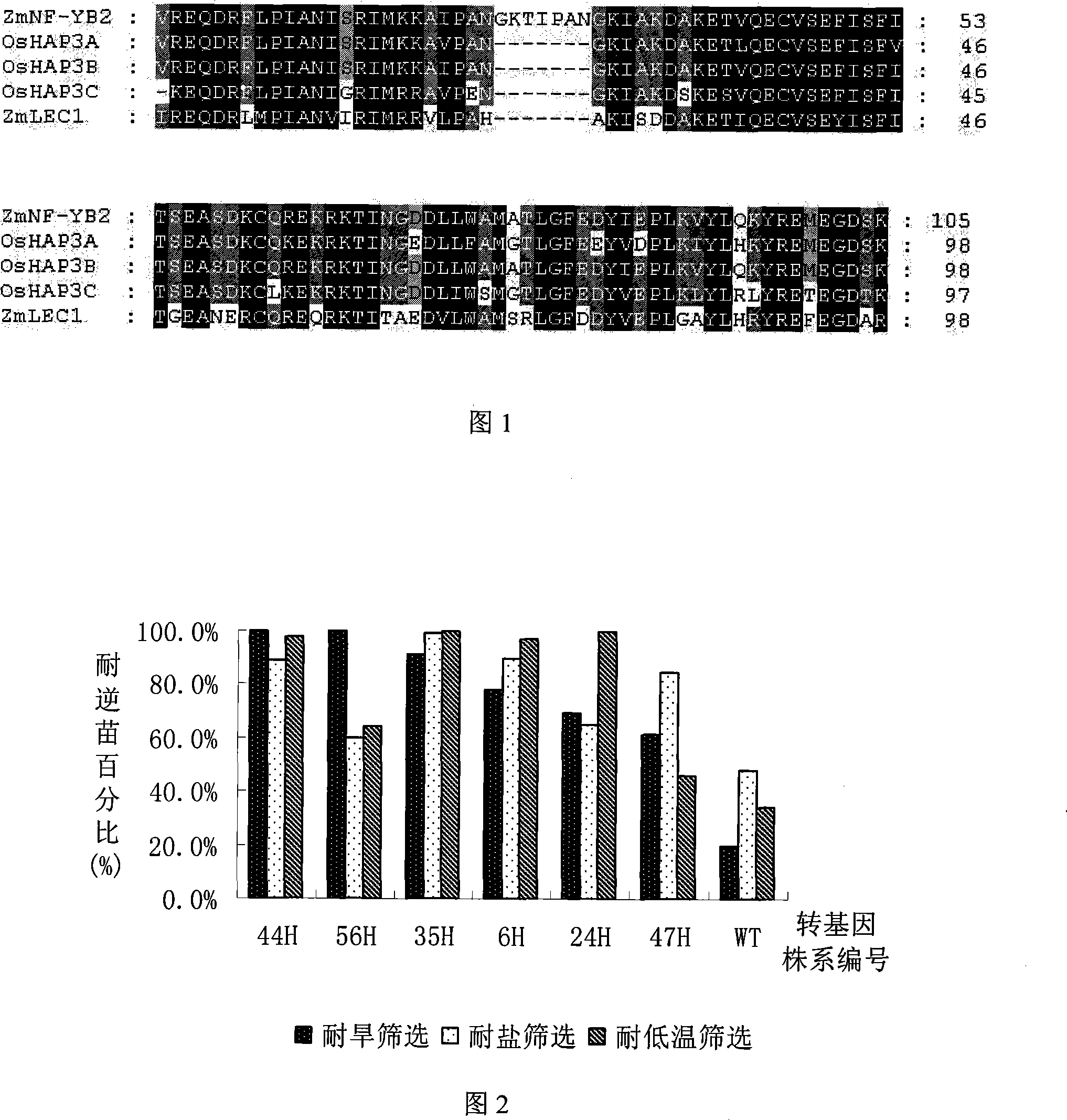
[0037] Example 1. Acquisition of genes encoding HAP transcription factors related to stress tolerance in rice
[0038] 1. Acquisition of gene sequence
[0039] OsHAP3A (GenBank No.: AB095438), OsHAP3B (GenBank No.: AB095439), and OsHAP3C (GenBank No.: AB095440) were queried from NCBI. (Miyoshi K, Ito Y, Serizawa A, Kurata N. OsHAP3 genes regulate chloroplast biogenesis in rice. The Plant Journal (2003) 36:532-540).
[0040] 2. PCR amplification and sequence analysis of OsHAP3A, OsHAP3B and OsHAP3C
[0041] Primers were designed according to the sequence in GenBank, and the cDNA of Oryza sativa "Zhonghua NO.11" was used as a template to amplify OsHAP3A, OsHAP3B and OsHAP3C by RT-PCR. The primer sequences are as follows:
[0042] (1) Design primers according to the CDS sequence of OsHAP3A:
[0043] OsHAP3A primer sequence:
[0044] Forward primer: 5'>CACCATGATGATGATGGATCTAGGGTTT>3'
[0045] Reverse primer: 5'>GGTGTCCCCATTATGATACTGAG>3'
[0046] (2) Design primers according...
Embodiment 2
[0057] Example 2. Acquisition of transgenic rice with genes encoding HAP transcription factors related to stress tolerance
[0058] The HAP transcription factor coding genes OsHAP3A, OsHAP3B and OsHAP3C related to stress tolerance obtained in Example 1 were transformed into rice by the Agrobacterium-mediated method, and the specific methods were as follows:
[0059] 1) Construction and identification of transformation vector
[0060] Artificially synthesized 182bp fragment containing endonuclease sites such as Sac I, Hind III and Spe I, which has the nucleotide sequence of SEQ ID NO: 7 in the sequence table, and the fragment is inserted after double digestion with Sac I and Spe I The pH2GW7 vector (purchased from inVitrogen) was used to obtain an intermediate vector containing a Hind III restriction site, which was named pHUG-1. After enzyme digestion and sequencing identification, it was proved that the inserted fragment was correct, and the construction of pHUG-1 was correc...
Embodiment 4
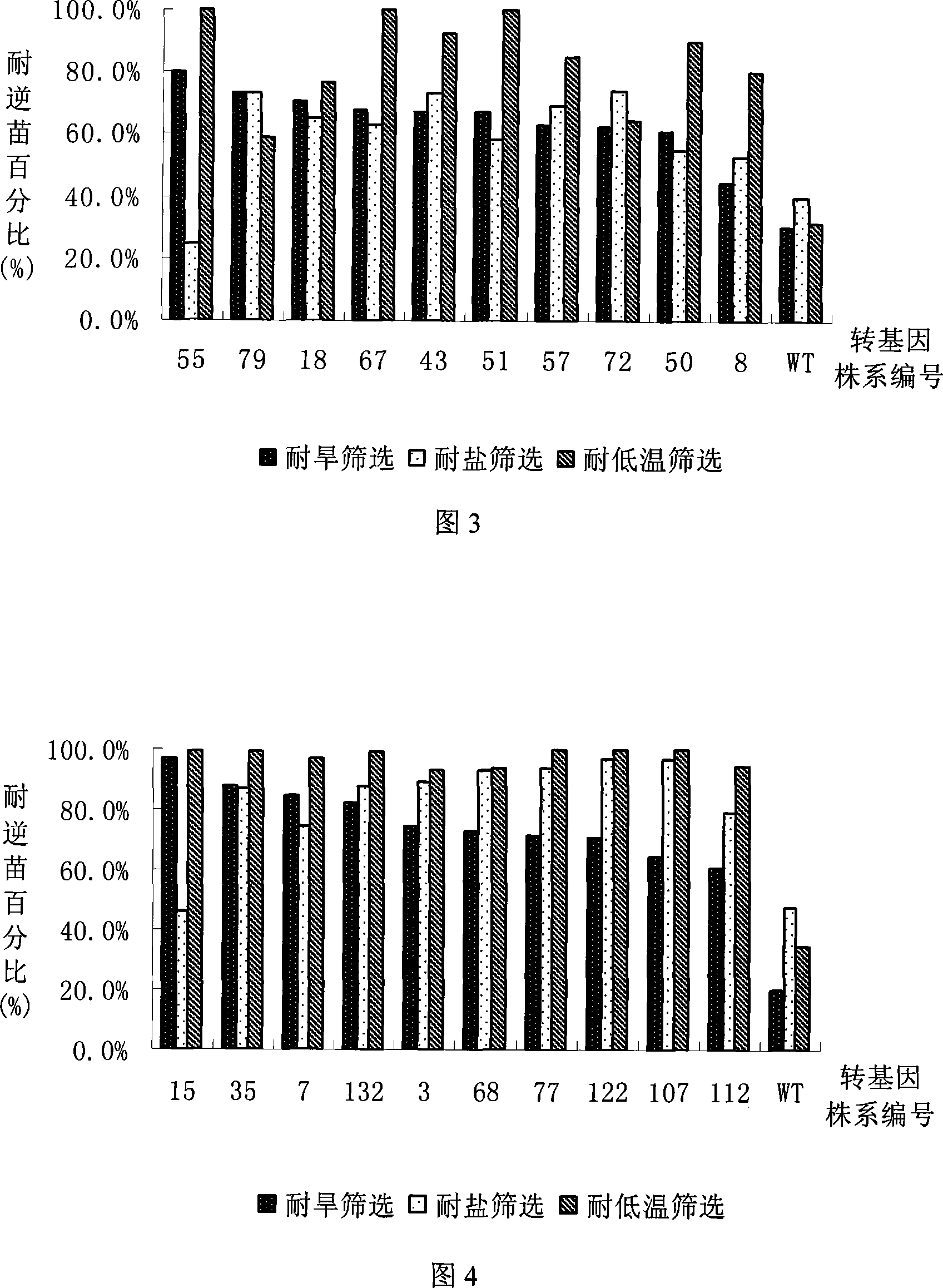
[0068] Embodiment 4, transgenic T 1 Drought Tolerance Identification of Progeny Plants
[0069] Harvest OsHAP3A, OsHAP3B and OsHAP3C T 1 Genetically modified rice seeds. Soak the seeds in water for 3 days to germinate them, then use 50 mg / L hygromycin for resistance screening, and use non-transgenic rice Zhonghua 11 as a control. After 5 days of screening, all the control plants died, and counted the resistant seedlings and dead seedlings of the transgenic plants To analyze the resistance segregation ratio of the transgenic plants, select the lines with a segregation ratio of about 3:1 between resistant seedlings and non-resistant seedlings. The results showed that OsHAP3A, OsHAP3B and OsHAP3C transgenic lines with single-site insertions were obtained. After continuing to cultivate for one week, drought treatment was carried out, and non-transgenic rice Zhonghua 11 plants were set as a control. Paddy rice seedling is planted in the soil of boxing, adopts vermiculite: the mi...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com