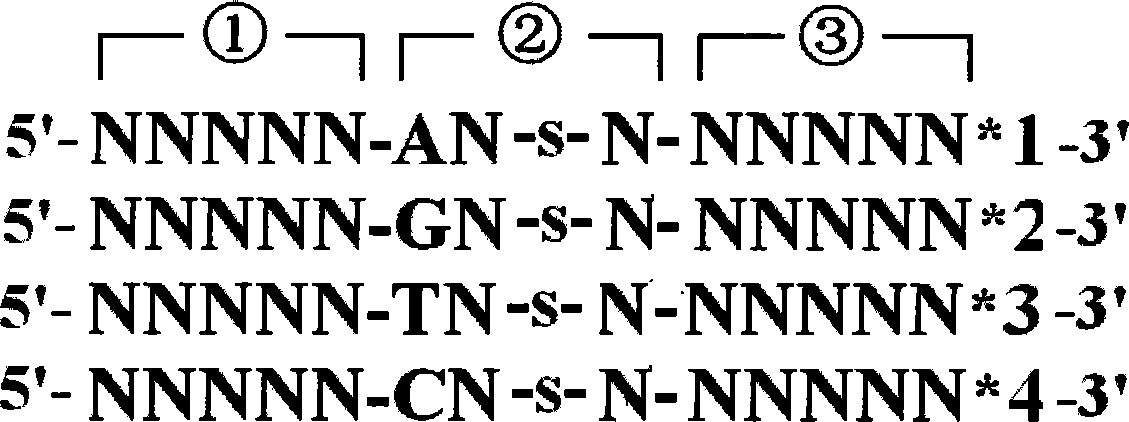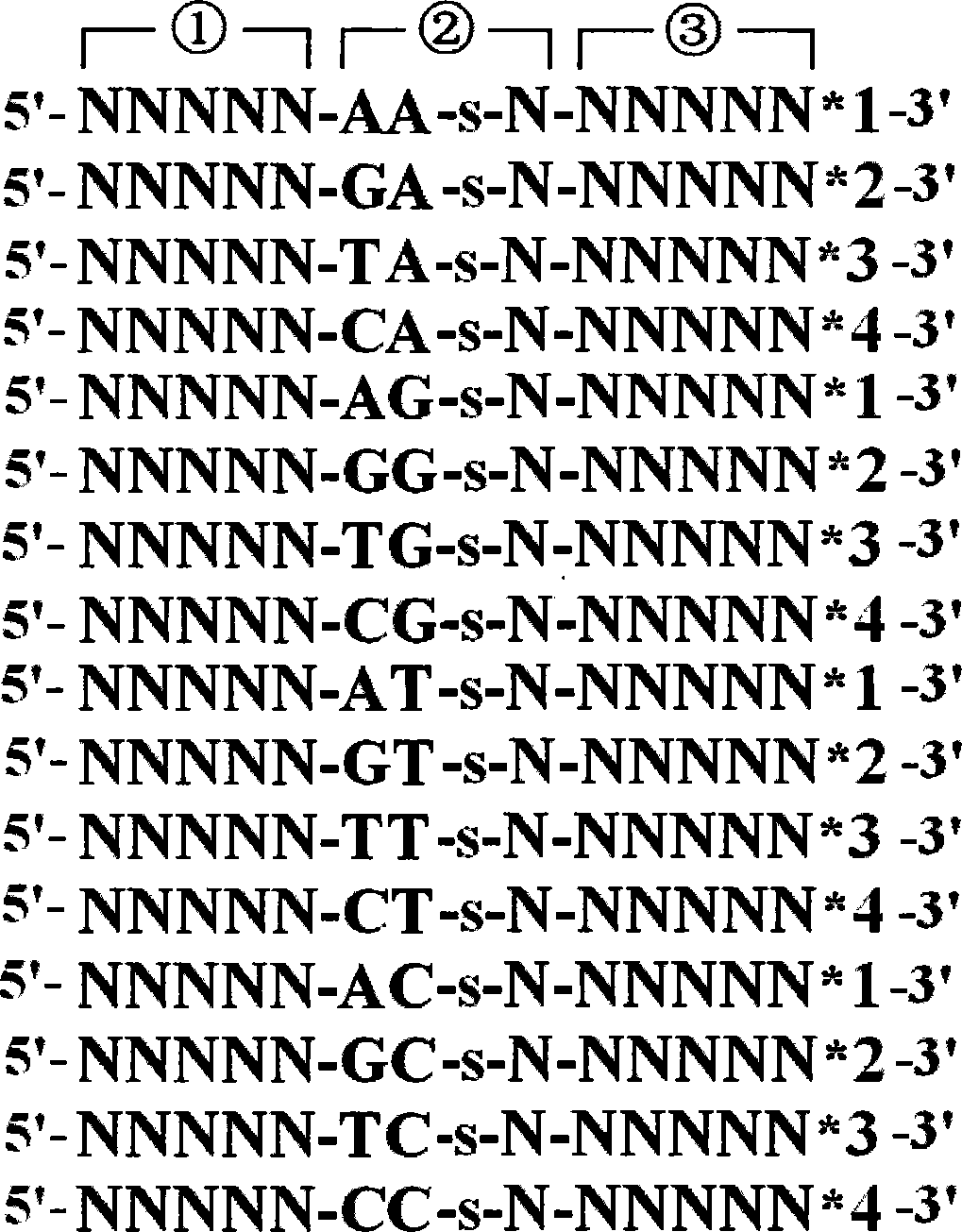DNA sequencing method by using thiooligonucleotide probe
A technology of oligonucleotide probes and thiooligonucleotides, which is applied in the field of DNA sequencing in biotechnology, can solve the problems of PCR amplification bias, easy introduction of replication errors, and short read length of synthetic sequencing methods, achieving Simple synthesis, easy implementation, accurate and reliable effect determination
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0021] Embodiment 1: Utilize the DNA sequencing method of thiooligonucleotide probe
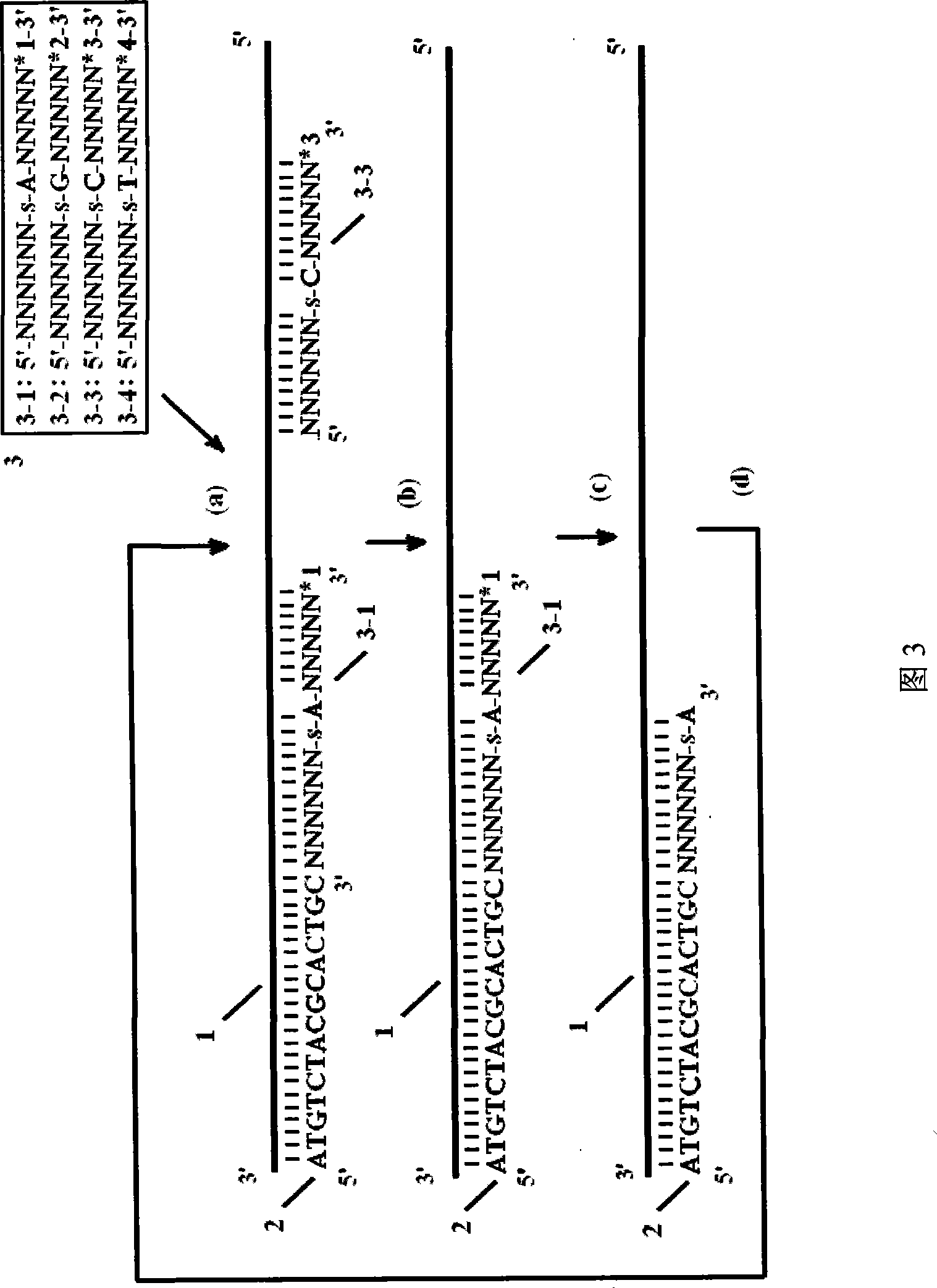
[0022] Using the DNA sequencing method of thiooligonucleotide probes, the sequencing steps are: construction of thiooligonucleotide probes: the 5'-3' end of the thiooligonucleotide probe sequence is the anchor region in turn , the recognition region and the splicing region, the anchor region contains n nucleotides or base analogs, where 0<n≤10; the recognition region contains m nucleotides or base analogs, where 0<m≤10 , and between the first nucleotide or base analog at the 3' end of the recognition region and the adjacent nucleotide or base analog in the 5' direction is a thio-modified phosphate bond; the cleavage region contains k nuclei Nucleic acid or base analogs, wherein 0<k≤10; the sequence of the shear region is provided with a marker corresponding to the recognition region; the sequencing cycle: a). Using sequencing positioning primers to hybridize with the single-stranded DNA templ...
Embodiment 2
[0023] Example 2: Determination of the whole human genome by single-base hybridization-ligation sequencing
[0024] Digest the human genome with enzymes (or sonicate) into fragments with a size of 50-200 bases, use T4 polymerase, kelnow polymerase, Tag polymerase and T4 phosphokinase to repair the ends of the fragments and form a phosphate group at the 5' end Under the action of T4 ligase, connect these fragmented nucleic acid sequences with a pair of universal linkers (5'-p-CAG TCA GTC AGT CAG TCA G T-3' and 3'-T GTC AGT CAGTCA GTC AGT C-p-5', where p represents a phosphate group) for connection, one oligonucleotide sequence of the universal linker is completely complementary to the sequence of the amplification primer, and the other oligonucleotide sequence of the universal linker is aligned with the sequencing Primers are the same.
[0025] The fragmented nucleic acid sequences connected by these linkers are subjected to emulsion parallel PCR reaction with microbeads immob...
Embodiment 3
[0027] Example 3: Determination of the whole genome of rice by two-base hybridization-ligation sequencing
[0028] Cut the rice genome with enzymes (or sonicate) into fragments with a size of 50-200 bases, use T4 polymerase, kelnow polymerase, Tag polymerase and T4 phosphokinase to repair the ends of the fragments and form a phosphate group at the 5' end Under the action of T4 ligase, connect these fragmented nucleic acid sequences with a pair of universal linkers (5'-p-CAG TCA GTC AGT CAG TCA G T-3' and 3'-T GTC AGT CAGTCA GTC AGT C-p-5', where p represents a phosphate group) for connection, one oligonucleotide sequence of the universal linker is completely complementary to the sequence of the amplification primer, and the other oligonucleotide sequence of the universal linker is aligned with the sequencing Primers are the same.
[0029] The fragmented nucleic acid sequences connected by these linkers are subjected to emulsion parallel PCR reaction with microbeads immobilizi...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - Generate Ideas
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com