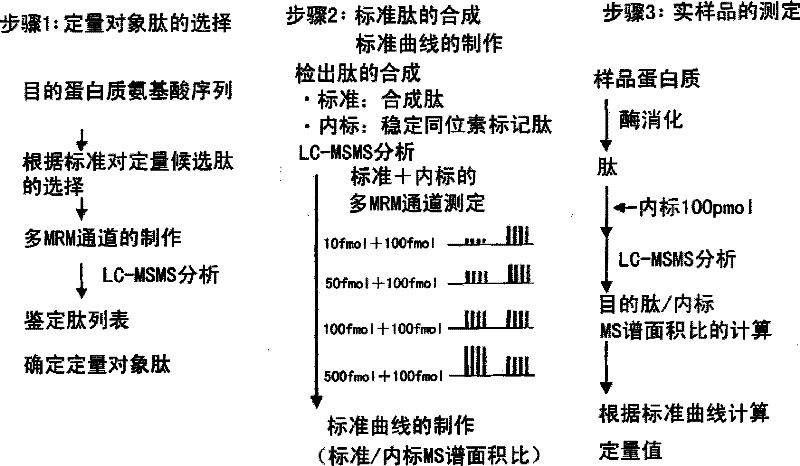
A Method for the Quantification of Membrane Proteins Using Mass Spectrometry
A technology for proteins and proteolytic enzymes, which is applied in the field of quantitative determination of membrane proteins existing in cell membranes, and can solve problems such as the reduction of quantitative accuracy.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0431] Peptide sample preparation
[0432] Human ABCG2 (human ATP binding cassette transporter G2) was selected as a cell membrane protein, and samples were prepared in order to select detectable proteins by LC-MSMS. First, using the cell membrane of ABCG2-expressing cells as a peptide supply source, it was prepared as follows. That is, the ABCG2 expressed cell membrane protein was denatured in a pH 8.5 buffer solution of 7M guanidine hydrochloride, 0.1MTris-HCl, and 10mM EDTA, and it was reduced with DTT in order to protect the SH group of the cysteine residue. and carbamidomethylation with iodoacetamide. Dialyze in 50 mM ammonium bicarbonate, add trypsin in an amount of 1 / 100 of the protein weight, and enzymatically digest at 37° C. for 16 hours to prepare peptide samples.
[0433] Preparation of quantitative target peptides and stable isotope-labeled peptides
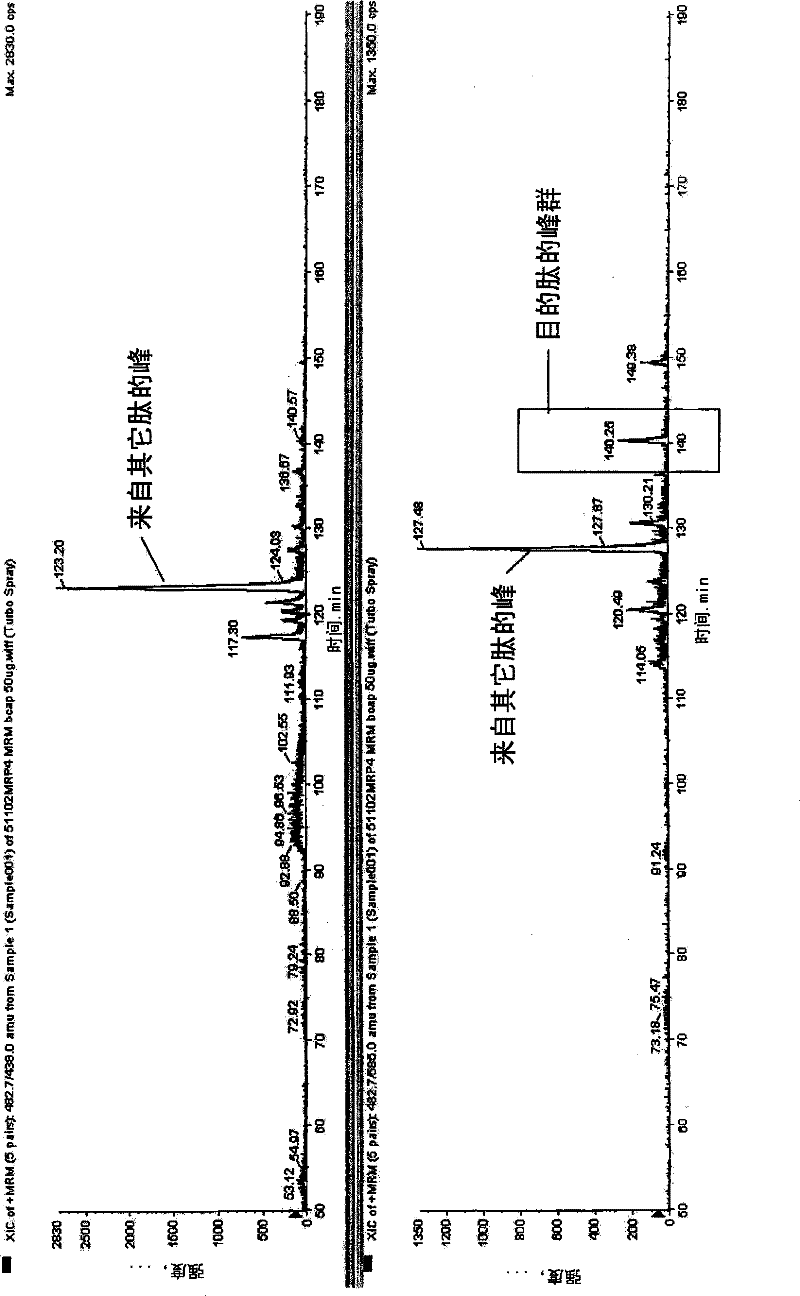
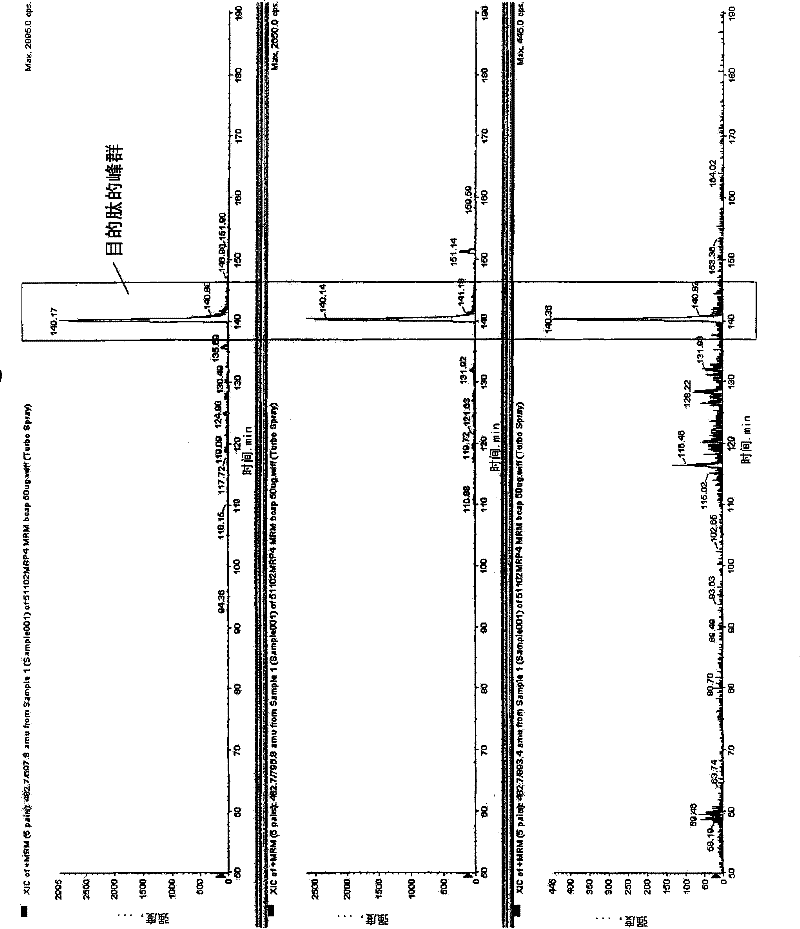
[0434] Human ABCG2 expressed cell membrane peptide samples were determined by LC-MSMS. From the measurement ...
Embodiment 2
[0441] As the criteria for selecting peptides to be quantified, (1), (2), (3), (4), (5), (6), (7), (8), (9) and (10) are used, and necessary When using (11), similarly to Example 1, a standard curve was prepared for the target peptides for protein synthesis and quantification described in Table 1 below, and its linearity was discussed. To 10 fmol, 50 fmol, 100 fmol, 500 fmol, and 1000 fmol of non-labeled peptides, 500 fmol of stable isotope-labeled peptide was added, respectively, and measured by LC-MSMS. Any of the above-mentioned peptides all show the linearity of the standard curve in the range of 10fmol-1000fmol, and it can be confirmed that the target protein can be quantified in this range ( Figure 6 ). In addition, as a transporter protein supply source, a cell membrane protein of a human leukemia cell line was selected for quantification. Table 1 shows the quantitative limit value of the obtained standard curve and the quantitative value of the leukemia cell membran...
Embodiment 3
[0446] As the criteria for selecting peptides to be quantified, (1), (2), (3), (4), (5), (6), (7), (8), (9) and (10) are used, and necessary When using (11), similarly to Example 1, for the proteins described in Table 2 below, peptides to be quantified were synthesized, a standard curve was prepared, and its linearity was discussed. 100 fmol of stable isotope-labeled peptide was added to 1 fmol, 5 fmol, 10 fmol, 50 fmol, 100 fmol, 500 fmol, and 1000 fmol of non-labeled peptide, respectively, and measured by LC-MSMS. Any of the above-mentioned peptides all showed the linearity of the standard curve, and it can be confirmed that the target protein can be quantified within this range ( Figure 6 ). Table 2 shows the quantitative limit value of the obtained standard curve and the quantitative value of the leukemia cell membrane.
[0447] Table 2
[0448] protein sequence stable isotope sequence Limit of quantitation (fmol) ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com