Recombinant plasmid for acetone-butanol clostridium gene disruption
A technology of Clostridium acetobutylicum and recombinant plasmid, applied in the field of genetic engineering, can solve the problems of inconvenient basic and applied research, imperfect genetic operating system, etc., and achieve the effect of improving insertion efficiency
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0046] Example 1 , Construction of pSY6 plasmid vector
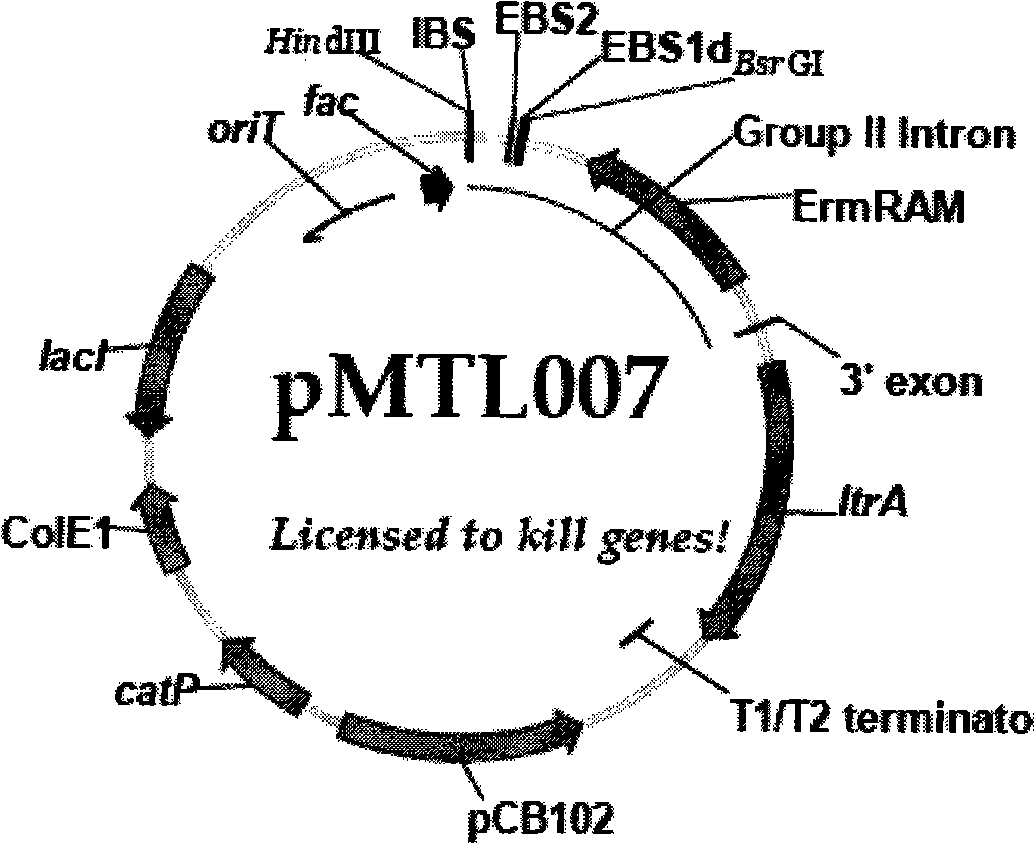
[0047] Using the pJIR750ai plasmid as a template, amplify the L1.LtrB class II intron and the IEP fragment (about 3.4kb in length) encoded by the ltrA gene contained in the plasmid by PCR, digest the amplified product with NdeI, and ligate it into The recombinant plasmid vector obtained from the E.coli-C.acetobutylicum shuttle vector pIMP1-ptb that was also digested and dephosphorylated was named pSY6. The specific experimental steps are as follows:
[0048] 1.1. Primer design and synthesis
[0049] Primer pair In-1 (SEQ ID NO: 4) and In-2 (SEQ ID NO: 5) were designed, NdeI and XhoI restriction sites were introduced at the 5' end of primer In-1, and restriction sites of NdeI and XhoI were introduced at the 5' end of primer In-2. An NdeI restriction site was introduced at the 5' end, where NdeI was used to construct pSY6, while XhoI facilitated the connection of subsequent targetron knockout fragments.
[0050] 1.2. P...
Embodiment 2
[0064] Example 2 , Construction of pSY6-buk, pSY6-solR plasmid vector
[0065] The buk targetron fragment and the solR targetron fragment were respectively amplified by PCR, digested with XhoI and BsrG I, and respectively ligated with the same digested pSY6 vector to obtain knockout plasmids pSY6-buk and pSY6-solR. Among them, the template and primer design of amplifying buk targetron and solR targetron are derived from the Targetron of Sigma-Aldrich Company TM Gene Knockout System (TA0100) Kit. Specific steps are as follows:
[0066] 2.1. Primer design and synthesis
[0067] ReferenceTargetron TM The method provided by the Gene Knockout System (TA0100) Kit designed primers buk-IBS (SEQ ID NO: 6), buk-EBS1d (SEQ ID NO: 7) and buk-EBS2 (SEQ ID NO: 8) respectively for constructing pSY6-buk plasmid vector; and solR-IBS (SEQ ID NO: 9), solR-EBS1d (SEQ ID NO: 10) and solR-EBS2 (SEQ ID NO: 11) were used to construct pSY6-solR plasmid vector.
[0068] In addition, it is also ...
Embodiment 3
[0083] Example 3 , Construct the recombinant strain Clostridium acetobutylicum of buk gene knockout
[0084] After the pSY6-buk plasmid was methylated at the Cac8 I site by E.coli ER2275 / pANS, it was electroporated into Clostridiumacetobutylicum ATCC 824. After recovery overnight, 100 μl of the cell solution was spread on the Amp (25 μg / ml) resistance plate, and the ℃, after 48-96 hours of cultivation in the anaerobic box, the colony PCR is used to verify the single bacteria, and the specific steps are as follows:
[0085] 3.1. Methylation of pSY6-buk plasmid
[0086] E.coli ER2275 was transformed with pANS to obtain E.coli ER2275 / pANS.
[0087] Prepare chemically competent cells of E.coli ER2275 / pANS, and then transform the pSY6-buk plasmid into E.coliER2275 / pANS. Since the pANS plasmid has spectinomycin resistance, it contains 100μg / ml Amp, 50μg / ml spectinomycin Cultivate overnight on the LB medium plate, and then pick a single colony to 4ml LB medium added with 100 μg / m...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com