Reporting bacterial strain sensitive to oxidation-reduction cycle reactant and preparation method thereof
A cycle reaction and bacterial strain technology, applied in biochemical equipment and methods, methods based on microorganisms, bacteria, etc., can solve the problems of large subjective errors, low accuracy, time-consuming and labor-consuming oxidation-reduction cycle reagents, etc., to achieve Easy to construct carrier, high sensitivity effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0055] Example 1. Fusion of target gene
[0056] (1) Primer information and synthesis
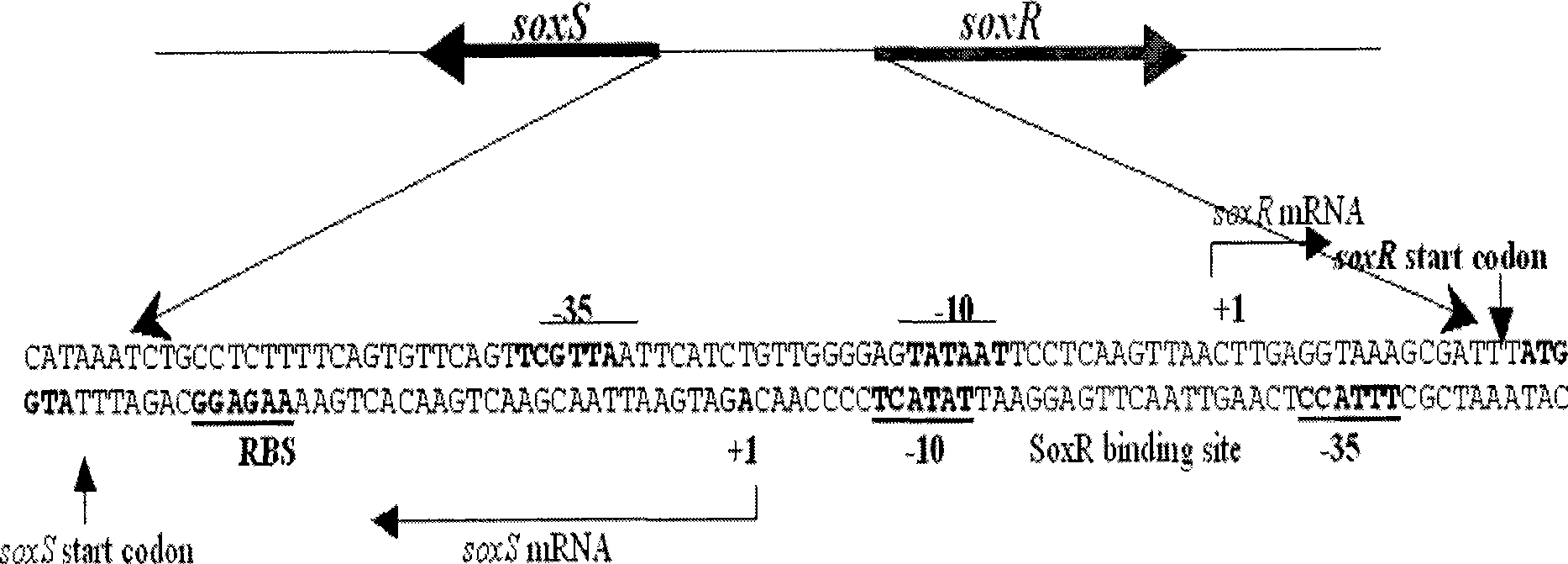
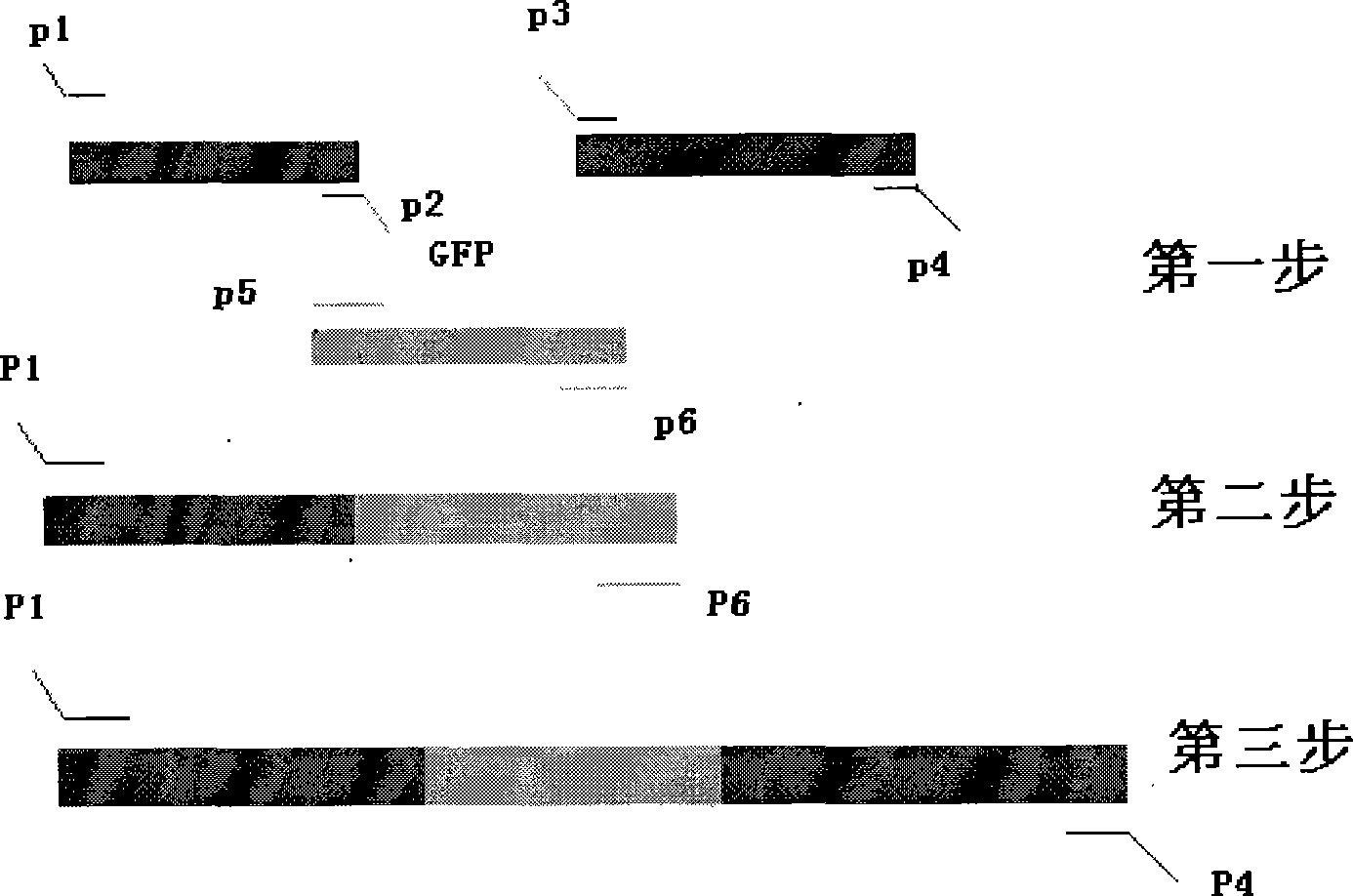
[0057] According to the gene knockout primer sequences provided by the Harvard University website and the SoxS and GFPmut2 gene sequences published by GenBank, the inner primers (P2:soxS-Ni and P3:soxS-Ci) and outer primers (P1 :soxS-No and P4:soxS-Co), so that there is a complementary sequence between P2 and P5, P3 and P6, the two outer primers for gene knockout remain unchanged, and P5 and P6 are a pair of primers for amplifying the GFPmut2 gene. The specific information is shown in Table-1. The primers were synthesized by Dalian Bao Biology Co., Ltd. The primers were dissolved in sterile deionized water to make a concentration of 10 μmol / L.
[0058] Table 1
[0059]
[0060] (2) Cross PCR for gene fusion
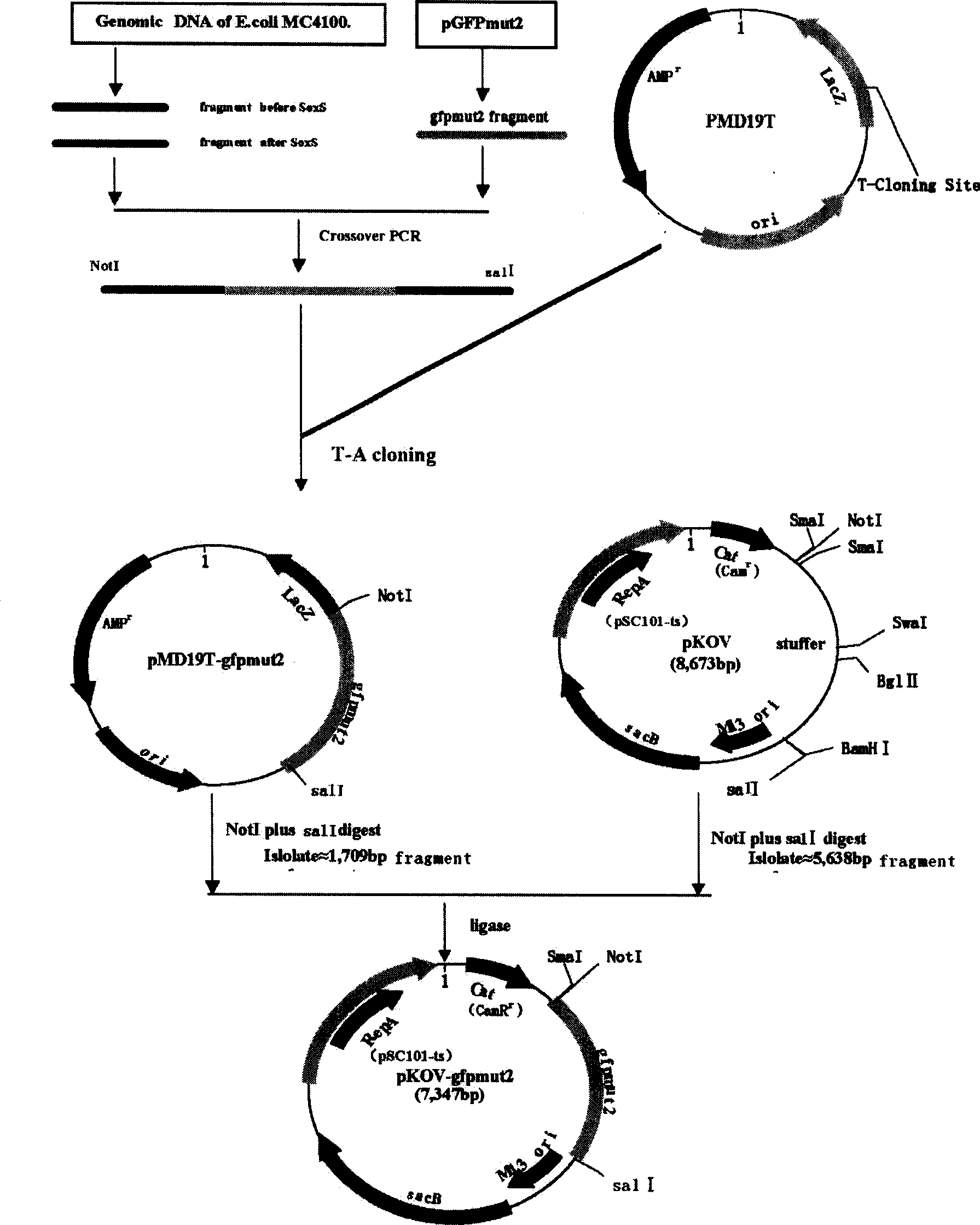
[0061] Using cross-PCR technology to carry out gene fusion through three-step PCR, the two-terminal sequence of the target gene SoxS is fused with the GFPmut2 gene to form a mod...
Embodiment 2
[0062] Example 2. Amplification of two homology arms and the GFPmut2 gene
[0063] 1. Amplification of two homology arms:
[0064] Use an inoculation needle to pick a small amount of a single colony of E.coli MC4100 on the LB plate, mix it in a 200 μL PCR reaction tube containing 15 μL sterile water, cook in boiling water for 10 minutes, and centrifuge at 12 000 rpm for 3 to 5 minutes to remove the water droplets condensed on the lid from the tube. end. Add 5 μL of the system prepared in Table 2 into the tube and mix well.
[0065] Table 2
[0066]
[0067] The PCR reaction conditions were as follows: 1 min at 94°C; 4 min at 88°C; 10 sec at 94°C, 3 min at 66°C, 25 cycles; 10 min at 72°C, and store at 4°C. 5 μL of the product was identified by 1.0% Agarose gel electrophoresis. The PCR product was purified with a PCR product purification kit, and finally dissolved and eluted by adding an appropriate amount of ddH2O for later use.
[0068] 2. Amplification of GFPmut2 gene: ...
Embodiment 3
[0080] Embodiment 3, PCR product adds A reaction
[0081] Add Ex Taq to the above PCR reaction product at a ratio of 5 U per 50 μL Polymerase, 72°C 10min.
[0082] (4), PCR fragment purification and recovery
[0083] Add A reaction product to purify with a PCR product purification kit, and finally add an appropriate amount of ddH2O to dissolve and elute for later use.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com