Method for identifying Rhizoma Corydalis Repentis
A Corydal Corydalis, a specific technology, applied in the field of identification of whole-leaf Corydalis Corydalis, can solve the problems that have not yet been seen such as rapid identification of the authentic DNA of Corydalis Corydalis, and achieve the effect of solving the problem of mixing and using less samples
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0040] Example 1: Preparation of whole-leaf Corydalis specific nucleotide sequence
[0041] 1. Genomic DNA Extraction
[0042] Leaf genomic DNA was extracted using the modified CTAB method (Doyle, 1991). Proceed as follows:
[0043] (1) Take about 0.05 grams of silica gel dried leaves, add a small amount of PVP powder, put it into the sample grinding tube, and grind it with BIO101 sample grinding machine, SPEED 4.0, TIME 20s.
[0044] (2) Quickly add 800-1000ul of 2×CTAB extraction buffer (containing 2% b-mercaptoethanol) preheated at 65°C, shake well and transfer to a 5ml centrifuge tube, then wash the grinding sample with 800-1000ul 2×CTAB tube, transferred to the same 5ml tube, 65. C water bath for 30 minutes.
[0045] (3) Cool to room temperature, add an equal volume of chloroform: isoamyl alcohol (24:1), mix well (slowly invert) for 15-30 minutes, and centrifuge at room temperature at 10,000 rpm for 10-15 minutes.
[0046] (4) Aspirate the upper aqueous phase, add 1 / 10...
Embodiment 2
[0060] Example 2: Preparation of whole-leaf Corydalis specific nucleic acid molecular probes Repens31 and Repens32
[0061] On the basis of obtaining the specific nucleotide sequence of the whole leaf Corydalis, utilize Primer Primer 5.0 (Vinay Singh, 1998) software to design, draw Repens31 and Repens32 (respectively 18-40bp shown in 1 in the sequence listing and The nucleotide composition and arrangement of 476-495bp) is a good oligonucleotide fragment for identifying the whole leaf Corydalis. According to the arrangement of the nucleotide composition of Repens31 and Repens32 (the nucleotide composition and arrangement shown in the 2 and 3 sequences in the sequence listing), it was synthesized on an automatic DNA synthesizer.
Embodiment 3
[0062] Embodiment 3: the identification of whole leaf Corydalis Corydalis (conventional PCR method)
[0063] 1. Extraction of DNA: The modified CTAB method was used to extract the total plant DNA.
[0064] 2. The PCR reaction system is:
[0065]
[0066] 3. PCR operation: take two 0.5 ml PCR tubes, add 22.5 microliters of PCR mixture according to step 2, then add 2.5 microliters of DNA to one tube, and 2.5 microliters of PCR mixture (control) to the other tube, and put On the PCR instrument, carry out the PCR reaction according to the following procedures:
[0067]
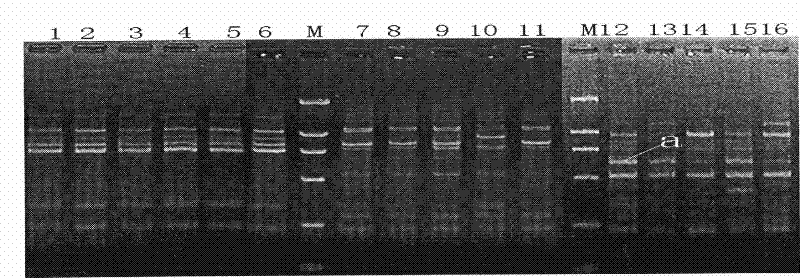
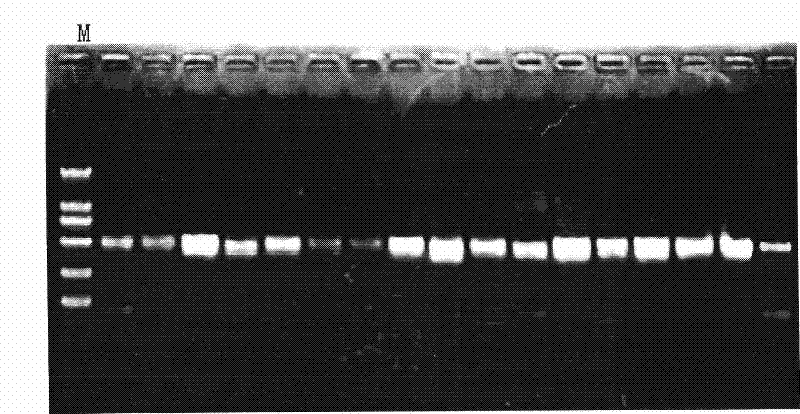
[0068] The results of PCR amplification were detected by electrophoresis on 1.5% agarose gel containing 0.1% EB. For electrophoresis results see figure 2 and image 3 . figure 2 It shows that there are bands around the molecular weight of 600bp in the whole leaf of Corydalis Corydalis, image 3 It shows that Corydalis has no bands around the molecular weight of 600bp. It shows that if Repens31 and R...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com