Double-plasmid expression system capable of producing virus-like particles containing large segments of RNA
An RNA virus and expression system technology, applied in the field of double plasmid expression system, can solve the problems of RNase sensitivity and lack of mature characteristics, and achieve the effect of reducing operating procedures and saving costs
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
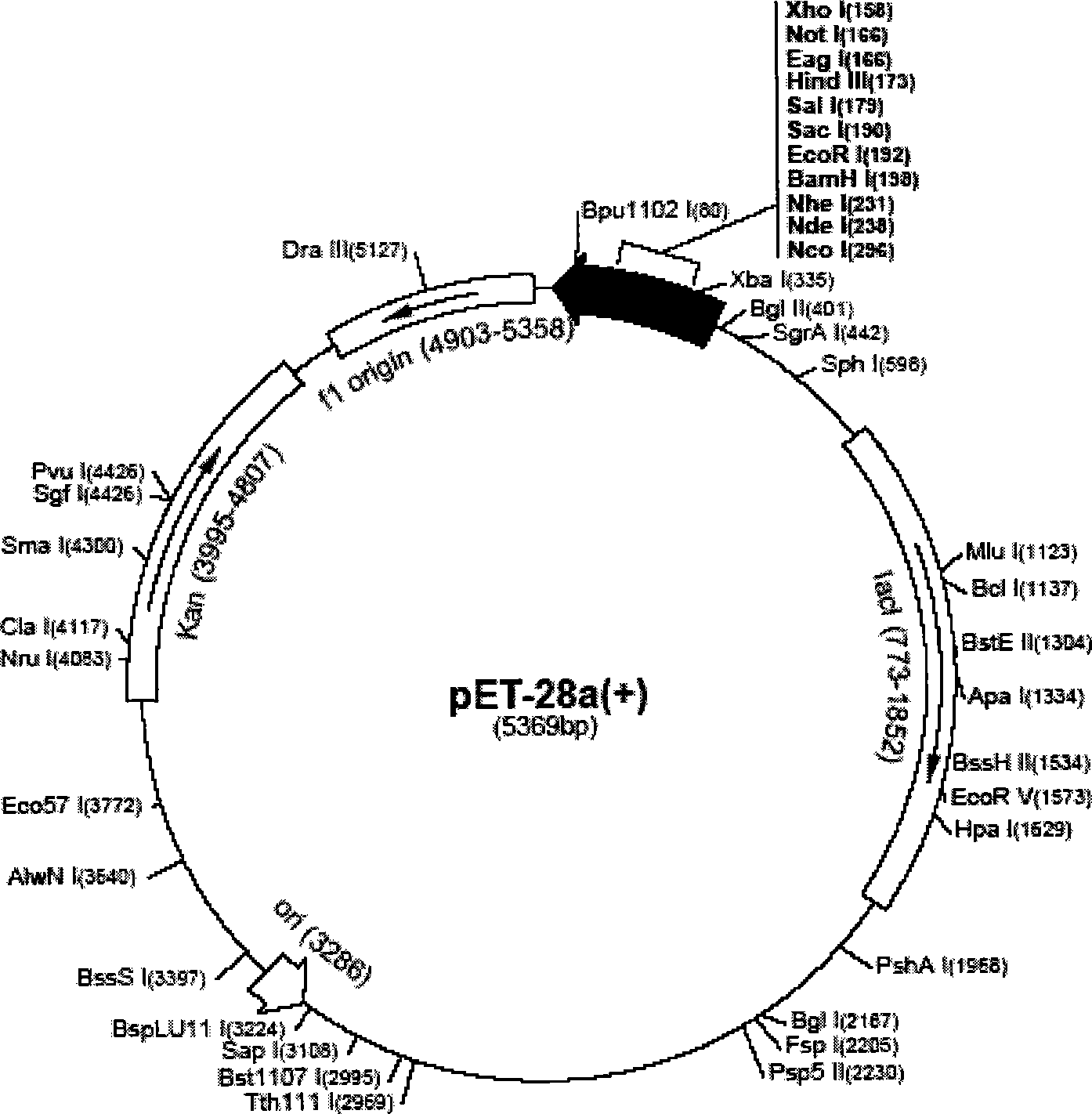
[0035] Example 1: Construction of pET-MC
[0036] 1. Amplification of the mature enzyme protein and capsid protein sequences of MS2
[0037] Using the pNCCL plasmid as a template, the mature enzyme protein and capsid protein sequence 81-1741 of MS2 were amplified by PCR.
[0038] Primer: 5'-CG G GATCCT GGCTATCGCTGTAGGTAGCC-3'81-101
[0039] 5'-CCC A AGCTT ATGGCCGGCGTCTATTAGTAG-3'1721-1741
[0040] Reaction system: 50μl each of 4 amplification tubes: 10*buffer5ul; Taq 0.4ul; DNTP 1ul; Primer11ul; Primer2 1ul; Mgcl 2 3ul; NCCL 1ul; DEPC water 37.6ul.
[0041] Cycle parameters: 94°C for 3 minutes; 94°C for 30 seconds, 57°C for 30 seconds, 72°C for 30 seconds, repeat 40 cycles; 72°C for 2 minutes; 72°C for 10 minutes.
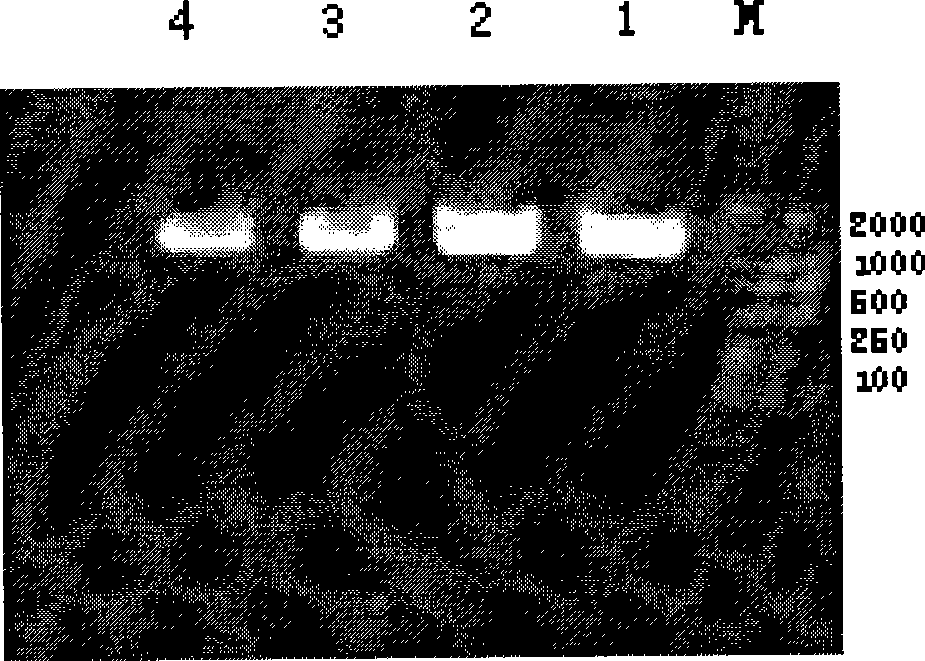
[0042] The results of electrophoresis showed that the length of the amplified fragment was 1700bp, which was in line with expectations.
[0043] 2. TA cloning of mature enzyme protein and capsid protein fragment of MS2
[0044] The MS2 mature...
Embodiment 2
[0052] Example 2: Construction of pACYC-3V
[0053] 1. Application of OVERLAP technology to amplify six chimeric sequences
[0054] The six sequences include three pieces of SARS-Cov (SARS-CoV1, SARS-CoV2, SARS-CoV3), one piece of HCV and two pieces of H5N1 (M300 and HA300).
[0055] The first round of PCR first amplifies six short sequences.
[0056] Amplification of SARS-CoV1: Template pBSSR-V6, donated by Peking Union Medical College Foundation. amplify
[0057] 15224-15618 sequence of SARS-CoV
[0058] Primers: S-SARS1: 5'TATCCAAAATGTGACAGAGCCATG3'LAP-SARS1: 5'ACGCTGAGGTGTGTAGGT GCAGGTAAGCGTAAAACTCATCCAC3'
[0059] Amplification of SARS-CoV2: Template pNCCL-SARS, constructed by our laboratory. Amplified SARS-CoV18038-18340 sequence
[0060] Primer: A-SARS2: 5'TAACCAGTCGGTACAGCTACTAAG3'
[0061] LAP-SARS: 5'AGTTTTACGCTTACCTGCACCTACACACCTCAGCGTTGATATAAAG3'
[0062] Amplification of SARS-CoV3: Template pBSSR7-8 was kindly provided by Peking Union Medical College Found...
Embodiment 3
[0098] Example 3: Transformation of double expression plasmids pACYC-3V and PET-MC into Escherichia coli BL21
[0099] 1. Co-transfection:
[0100] The two plasmids were mixed in equal volumes for a total of 10 ul, and 10 ul of the plasmid was added to a tube containing 200 ul of competent cells, gently swirled several times to mix the contents, and placed in ice for 30 minutes. Place the tube in a circulating water bath pre-warmed to 42 °C for 90 s without shaking. Quickly remove the tube to an ice bath and allow the cells to cool for 2 min. Add 200ul of non-resistant LB medium. Incubate with shaking at 37°C for 1 hour. Then spread on plates containing chloramphenicol and KANAmycin. Leave the plate at room temperature until the liquid has absorbed. Invert the plate and incubate at 37°C for 16 hours. As a result, colony growth was visible in the culture medium, which was in line with the expected results.
[0101] 2. Extract the plasmid:
[0102] Pick the transformed s...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com