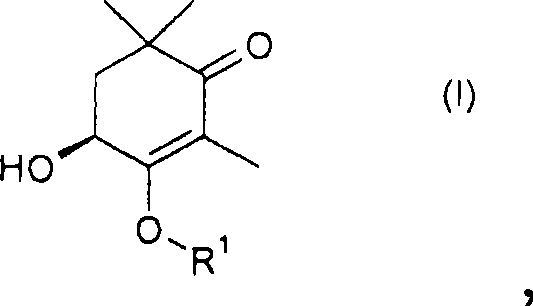
Method for the production of (4s)-3, 4-dihydroxy-2,6, 6-trimethyl-cyclohex-2-enone and derivatives thereof
A technology of derivatives and hydroxyl, which is applied to the preparation of (4S)-3 by using the phenylethanol dehydrogenase of Azotoxabacterium, which can solve the problems of unsuitable industrial scale implementation and uneconomical
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0088] Embodiment 1: Preparation of recombinant phenyl alcohol dehydrogenase
[0089] In a 100ml Erlenmeyer flask (baffle), Escherichia coli (E.coli) LU11558 prepared as described in WO 2005 / 108590 Examples 1 and 2 was dissolved in 20ml of LB-Amp / Spec / Cm (100 μg / l ampicillin Penicillin, 100 μg / l spectinomycin, 20 μg / l chloramphenicol), 0.1 mM IPTG, 0.5 g / L rhamnose, cultured at 37°C for 18 hours, and then cultured at 5000 * g for 10 min with 10 mM TRIS * Wash once with HCl (pH 7.0) and resuspend in 2 ml of the same buffer.
[0090] Cell-free crude protein extracts were prepared by disrupting E. coli LU11558 cell paste with 0.7 ml glass beads (d = 0.5 mm) in a vibrating ball mill (3 x 5 min, cooled on ice at intervals).
Embodiment 2
[0091] Example 2: Determination of the activity of the recombinant dehydrogenase obtained from Escherichia coli LU11558
[0092] 10 μl of cell-free crude extract (Example 1; approximately 10 mg / ml total protein) was shaken and incubated in 770 μl of 50 mM potassium phosphate buffer (with 1 mM MgCl 2 , pH 6.5), 100 μl of isopropanol, 100 μl of NADH solution (0.5M) and 20 μl of compound 1 (1M in DMSO). The mixture was analyzed analogously to Example 3. On average 0.13 mM of 3,4-dihydroxy-2,6,6-trimethylcyclohex-2-enone was formed. There was no detectable conversion in control experiments in which rhamnose was not added during incubation.
Embodiment 3
[0093] Embodiment 3: analysis compound 1 and compound 2
[0094] Precursor and product concentrations can be determined by HPLC. In addition to concentration, ee can also be determined based on the choice of stationary and mobile phases.
[0095] Stationary phase: Chiralpak AS-RH, 150*4.6mm, Daicel, equilibrated at 40°C
[0096] Mobile Phase: Eluent A: 10mM KH 2 PO 4
[0097] Eluent B: CH 3 CN
[0098] Gradient: Time [min] A[%] B[%] Flow rate [mL / min]
[0099] 0 90 10 0.5
[0100] 10 90 10 0.5
[0101] 11 60 40 0.5
[0102] 20 60 40 0.5
[0103] Flow rate: 0.5ml / min
[0104] Detection: UV detection at 260 nm
[0105] Dwell time:
[0106] (+)-3,4-dihydroxy-2,6,6-trimethylcyclohex-2-enone: about 9.3 minutes
[0107] (-)-3,4-dihydroxy-2,6,6-trimethylcyclohex-2-enone: about 9.8 minutes
[0108] 2-Hydroxy-3,5,5-trimethylcyclohex-2-ene-1,4-dione: about 17.6 minutes
[0109] Using authentic materi...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com