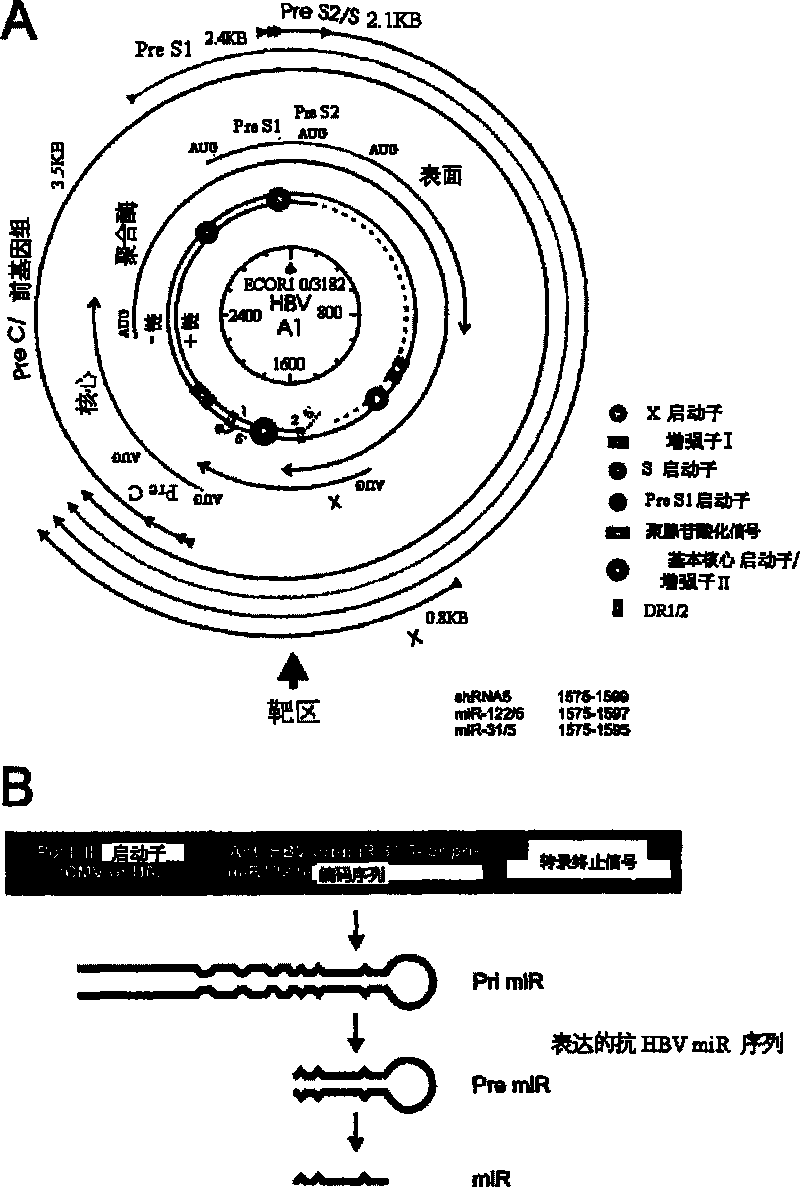
A primary micro RNA expression cassette
An expression cassette and primary technology, applied in the field of viral gene expression inhibition, can solve problems such as the inability to generate functional RNAi effectors
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0279] Example 1: Design and Proliferation of Anti-HBV pri-miR Expression Plasmids
[0280] 1. Design
[0281] By replacing naturally occurring miR-31 (SEQ ID NOs: 1 / 107 / 133) and miR-122 (SEQ ID NOs: 2 / 108 / 134) to design pri-miR expression cassettes. The complete sequence encoding the anti-HBVpri miR sequence is given in SEQ ID NO: 3(135) and 4(136). The wild-type sequences of miRs were maintained as much as possible and the computer-aided prediction of transcript secondary structure [38] would not be significantly different from that of the corresponding wild-type miRs. The final expression cassette contained 51 nucleotides of wild-type sequence flanked by either end of the pre-miR (Zeng and Cullen, 2005). In order to facilitate the cloning of miR, Nhe I and Spe I restriction enzyme sites were introduced at its 5' and 3' ends, respectively. figure 1 A schematic diagram of the target genomic loci of HBV together with the pri-miR expression cassette is shown in . figure ...
Embodiment 2
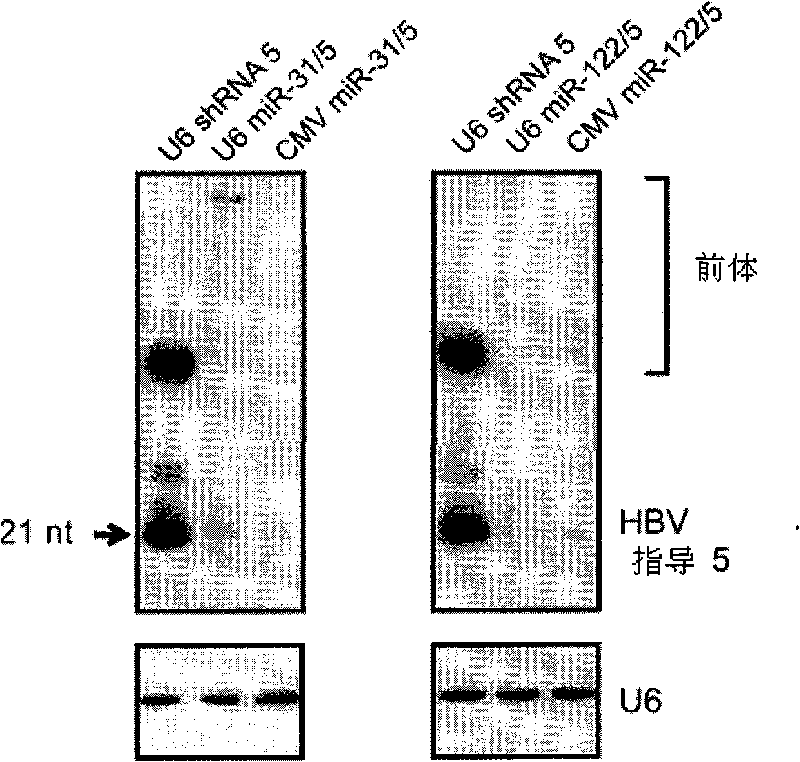
[0314] Example 2: Intracellular expression of anti-HBV sequences from pTZ-57R / Tpri-miR-31 / 5 and pTZ-57R / Tpri-miR-122 / 5 in transfected cultured cells.
[0315] Cultured cells were transfected with plasmids encoding miR-31 / 5 and miR122 / 5.
[0316] HEK293 cells were propagated in DMEM supplemented with 10% FCS, penicillin (50 IU / ml) and streptomycin (50 μg / ml) (Gibco BRL, UK). One day before transfection, 1,500,000 HEK293 cells were seeded into a petri dish with a diameter of 10 em. Transfection was performed with 10 μg of shRNA- or miR-expressing plasmids using cationic liposomes (Invitrogen, CA, USA) following the supplier's instructions.
[0317] Northern blot analysis. HEK293 cells were harvested 4 days after transfection, and total RNA was extracted using Tri reagent (Sigma, MI, USA) according to the supplier's instructions. 20 μg of RNA was resolved in a urine-denaturing 12.5% polyacrylamide gel and printed on a nylon membrane. Radiolabeled DNA oligonucleotides serve ...
Embodiment 3
[0319] Example 3: Anti-HBV Efficacy Test of miR Sequences in a Cell Culture Model of HBV Replication
[0320] target plasmid. pCH-9 / 3091 was described previously [42]. It contains HBV sequences larger than the length of the genome, which is similar to the HBV Al subgenotype consensus, and a 3.5 kb HBV pregenomic transcript is generated from its CMV promoter. The pCH firefly Luc vector was prepared by replacing the preS2 / SORF of pCH-9 / 3091 with firefly luciferase-encoding DNA following a procedure similar to that used to generate pCHGFP as described previously [43]. Briefly, the firefly luciferase sequence was amplified from psiCheck2 (Promega, WI, USA) by PCR. The primer combination for amplifying the coding region of the firefly luciferase sequence is:
[0321] 5' ACTGCTCGAGGATTGGGGACCCTGCGCTGAACATGGTGAGCAAGGGCG3' (forward; SEQ ID NO: 52);
[0322] and 5'ACGTTCTAGGTATACGGACCGTTACTTGTACAGCTC3' (reverse; SEQ ID NO: 53).
[0323] The forward primer contained a sequence comp...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com