Recombinant adenovirus vector for efficiently inducing pluripotent stem cell (PS cell), method for inducing PS cell by using recombinant adenovirus vector and usage of recombinant adenovirus vector
A technology of pluripotent stem cells and recombinant adenovirus, which is applied in the field of genetic engineering and stem cell science, can solve the problems of embryonic stem cell differentiation and the inability of embryos to form epiblasts
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
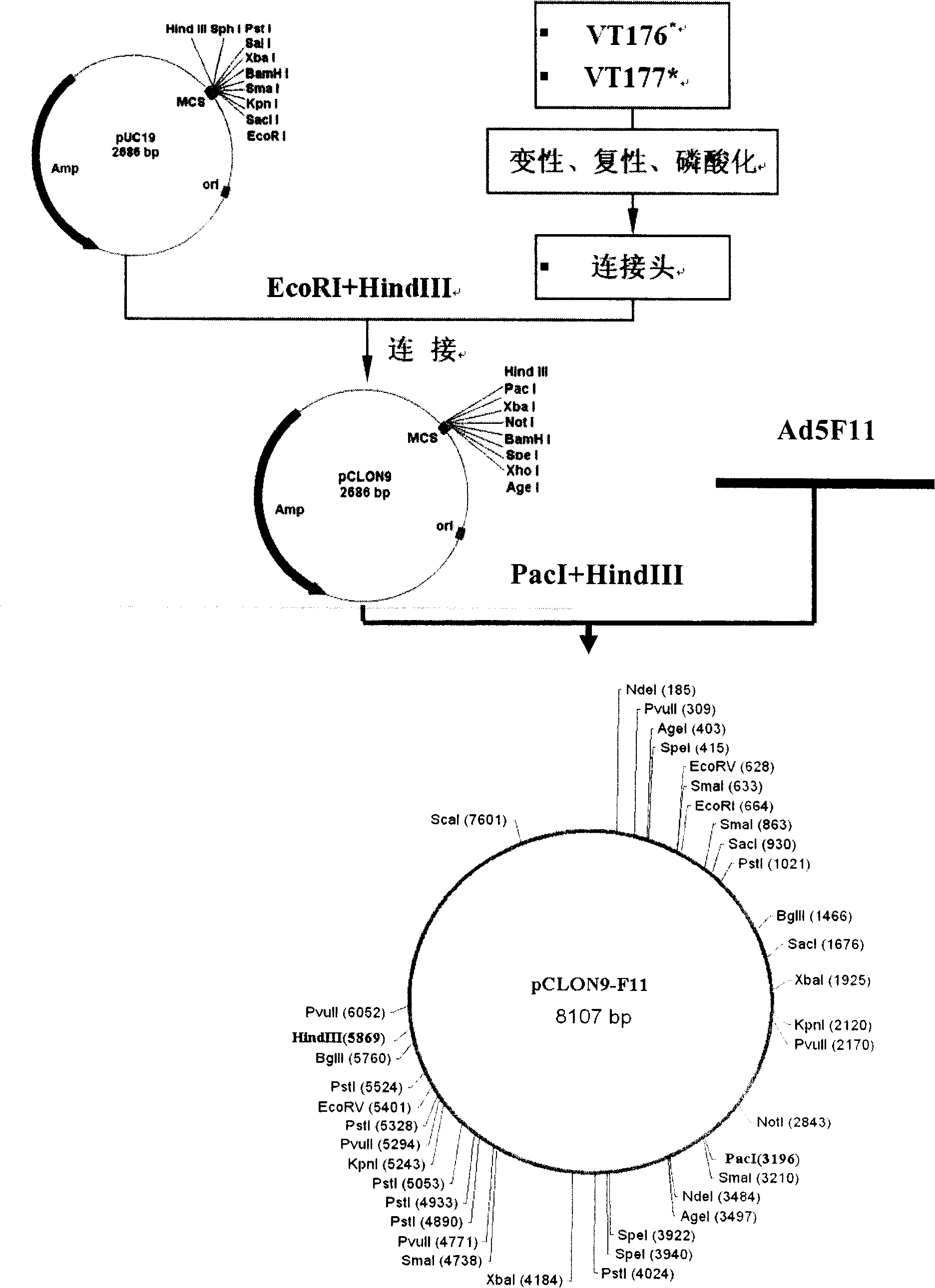
[0089] Example 1: Taking the type 5 adenovirus vector as an example, the construction of the adenovirus right-arm plasmid pPE3-F11 containing the head coding sequence of the Ad11 adenovirus cilium is illustrated
[0090] Step 1: Synthesize (TAKARA) adenovirus type 11 ciliary protein gene coding sequence (sequence source: NC_011212, GenBANK) as shown in SEQ ID NO: 3, named Ad5F11, wherein: (1) 232-365 bases : type 5 adenovirus ciliary protein tail sequence; (2) bases 364-1212: type 11 adenoviral ciliary protein shaft and knob sequences; (3) the rest of the bases: type 5 adenovirus genome sequence, see Microbix Biosystems pBHGloxdeltaE1, 3Cre sequence.
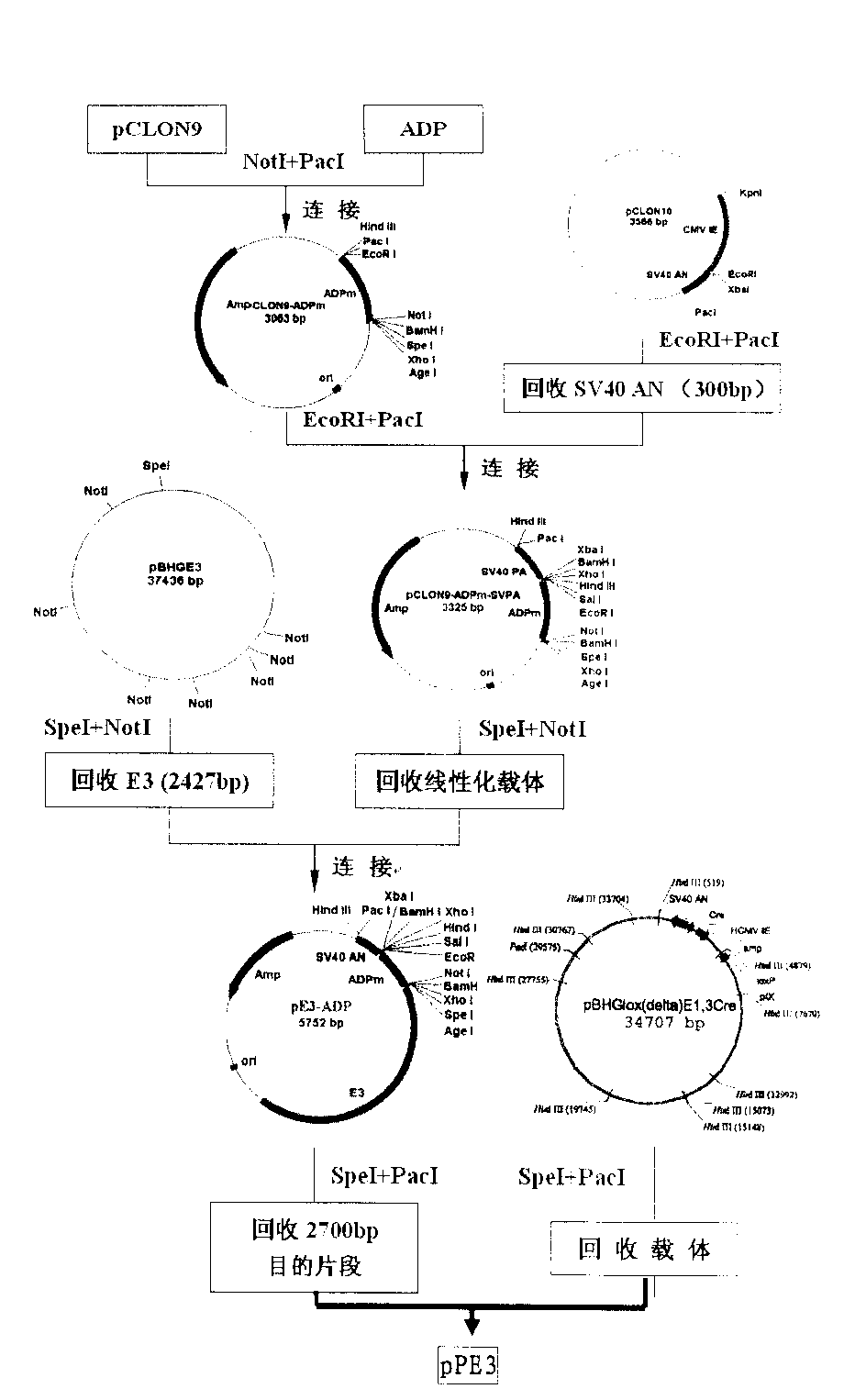
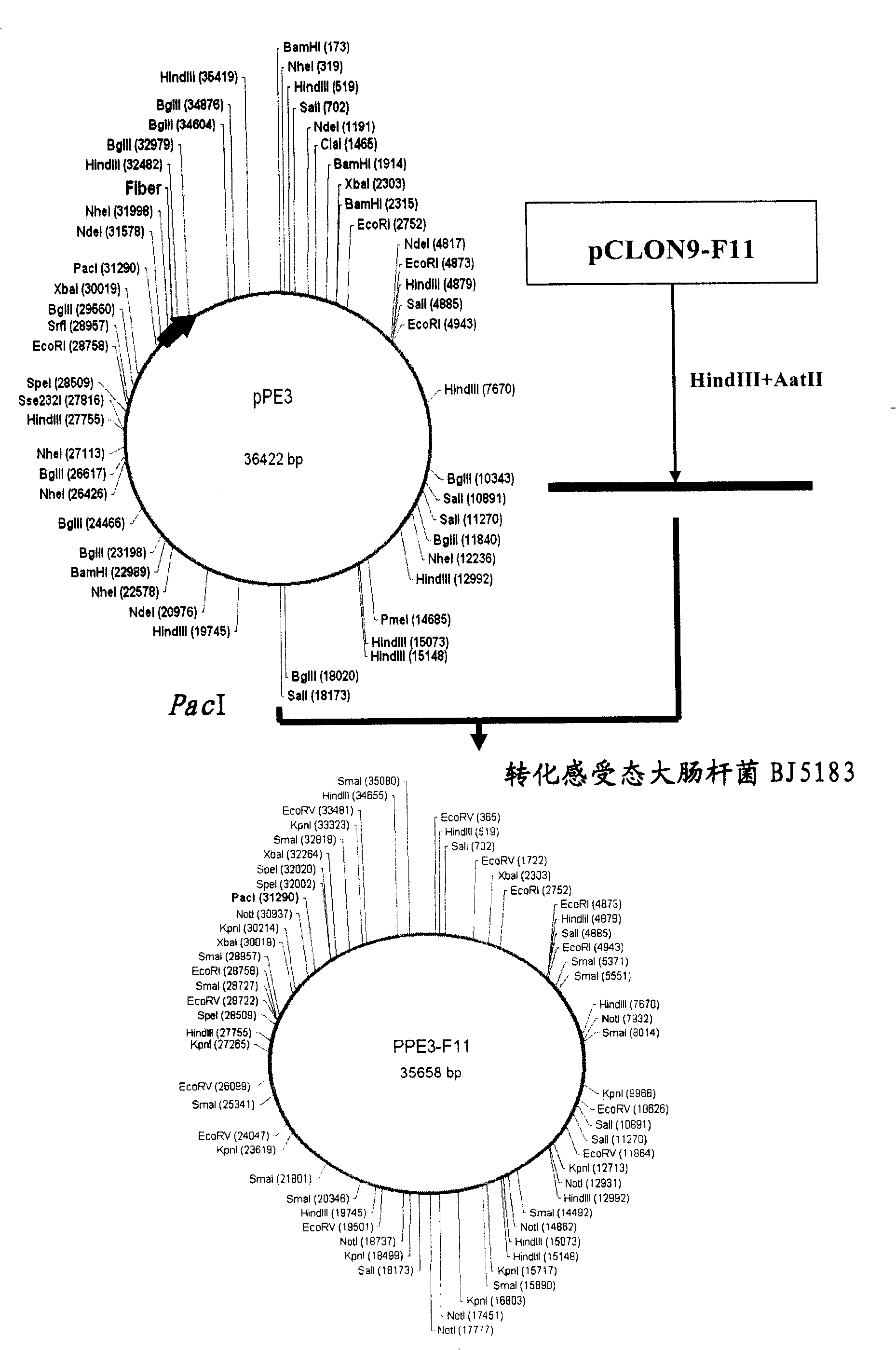
[0091] Step 2: Synthesize the VT176 sequence (SEQ ID NO: 4) and the VT177 sequence (SEQ ID NO: 5) to construct the pCLON9 vector. The Ad5F11 sequence was digested with PacI+HindIII, and a 2656bp fragment was recovered, which was ligated with the vector digested with pCLON9 / PacI+HindIII, confirmed by enzyme digestion and sequenc...
Embodiment 2
[0095] Example 2: Construction of pDC328-OSN and pDC328-OKS
[0096] Step 1: Synthesize the EF1α promoter sequence (TAKARA), see SEQ ID NO: 7 for the sequence.
[0097] Step 2: The synthesized EF1 was double digested with XbaI+SalI, and a 600bp fragment was recovered.
[0098] Step 3: The pDC315 vector (purchased from Microbix Biosystem Inc. (Toronto), containing the 1417 to 2344bp sequence fragment of the left arm E1 region of type 5 adenovirus, and the inverted terminal repeat sequence) was digested with XbaI+SalI double enzymes, and the 3345bp fragment was recovered .
[0099] Step 4: Ligate the fragments recovered in Step 3 and Step 4, and confirm by enzyme digestion and sequencing (Invitrogen). The correct one named the vector as pDC328.
[0100] Step 5: Synthesize OCT4-2A-SOX2-2A-NANOG sequence (TAKARA), sequence source: OCT4 NM_002701; SOX2 NM_033106; NANOG NM_024865, GenBank. The sequence is shown in SEQ ID NO: 8, wherein: (1) bases 14-1093 are the coding sequence o...
Embodiment 3
[0105] Example 3: Construction of pDC328-EGFP and pDC315-EGFP
[0106] Step 1: Synthesize EGFP sequence (TAKARA), see SEQ ID NO:10 for the sequence.
[0107] Step 2: The synthesized EGFP was digested with EcoRI+SalI to recover a 726bp fragment.
[0108] Step 3: pDC328 and pDC315 were digested with EcoRI+SalI to recover 3897bp and 3883bp fragments respectively.
[0109] Step 4: The fragments recovered in the second step were ligated with the fragments recovered by digestion with pDC328 and pDC315 respectively.
[0110] The construction process of pDC328-EGFP and pDC315-EGFP is as follows Figure 5 shown.
PUM
| Property | Measurement | Unit |
|---|---|---|
| diameter | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com