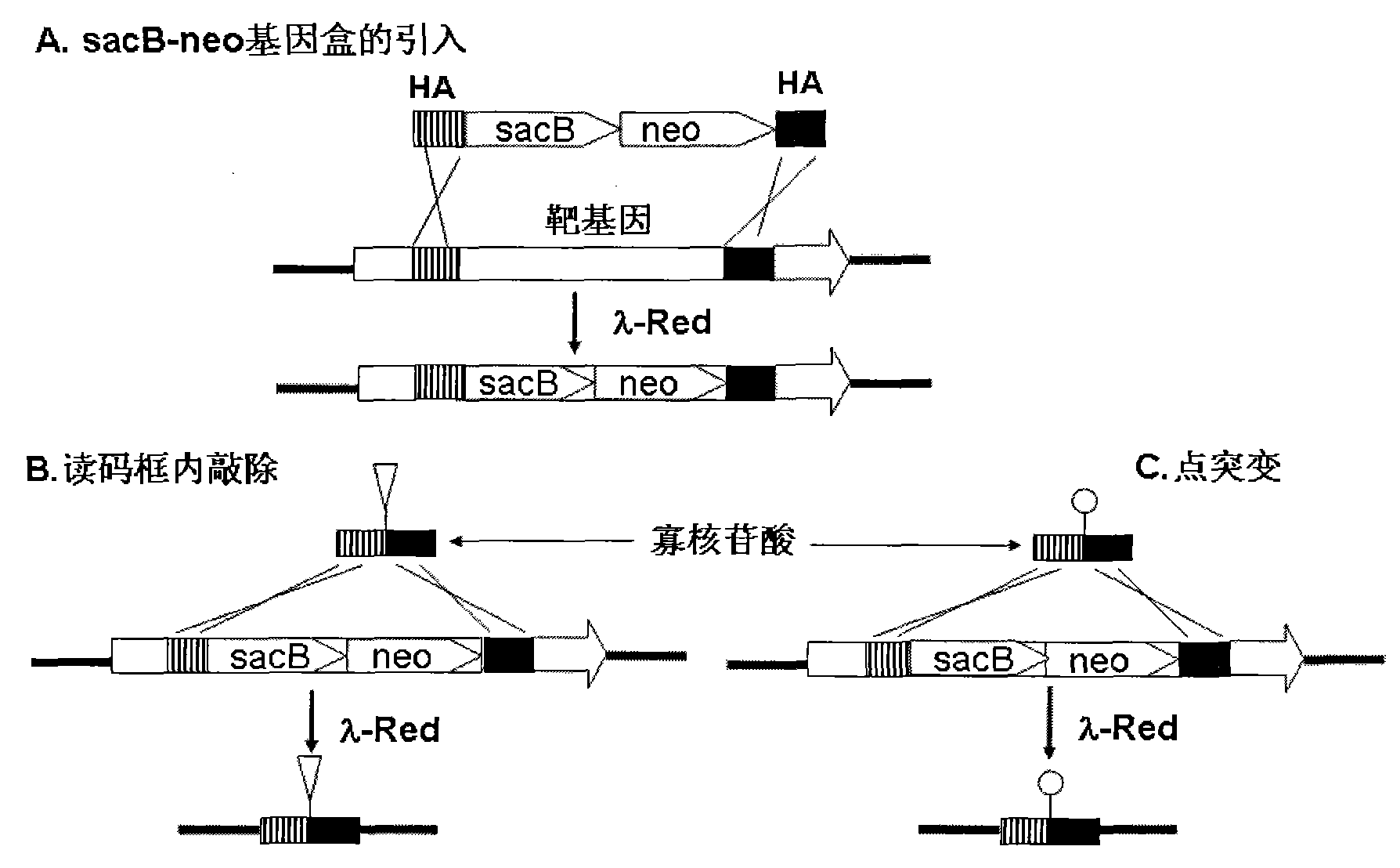
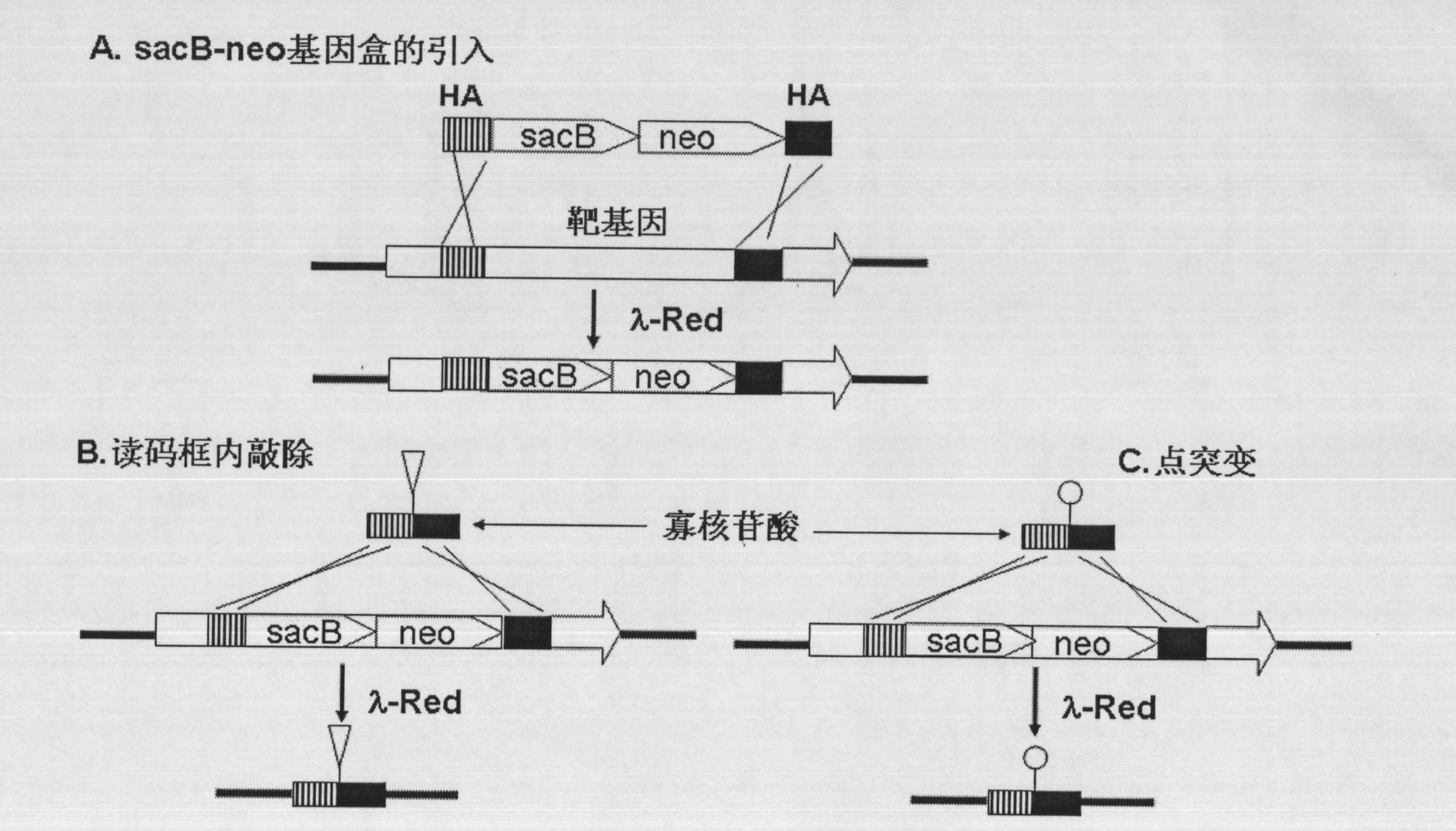
Oligonucleotide mediated colibacillary gene knock-out or point mutation method
An oligonucleotide and Escherichia coli technology, applied in the field of genetic engineering, can solve the problems of increasing time and difficulty, affecting other gene expression, interfering with the overall structure of the genome, etc., to avoid polarity effects, high recombination efficiency, and easy operation line effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0059] Embodiment 1. Construction of plasmid pSNR containing sacB-neo gene cassette
[0060] 1. Preparation and DNA transformation of Escherichia coli MG1655, DH10B and Escherichia coli BW25141 electroporation competent cells
[0061] Inoculate a freshly streaked single colony from the plate into 2ml LB liquid medium and shake overnight at 37°C, then transfer to 50ml LB at 1 / 50 volume, and shake until the cell OD 600 About 0.6. Pour the bacterial solution into a pre-cooled centrifuge tube, place in an ice bath for 10 minutes, centrifuge at 5000 rpm for 5 minutes at 4°C, and discard the supernatant. Wash the pellet twice with 10% glycerol, and finally suspend in 200 μl of 10% glycerol, and distribute in 50 μl per tube.
[0062] To prepare Escherichia coli competent cells containing the plasmid, corresponding antibiotics were added to the LB liquid medium during the culture of the strain.
[0063] Add the DNA to 50 μl of ice-thawed electroporation-competent cells, and mix well ...
Embodiment 2
[0080] Example 2. LacZ gene knockout
[0081] 1. Amplification of the sacB-neo gene cassette with homology arms
[0082] Design primer ZDE1: 5′- TGGCCGTCGTTTTACAACGTCG TGACTGGGAAAAC CCTGGCGTTACCCAA ATCCTTTTATGATTTTCTATC-3' (SEQ ID NO.3), the first 50 bases of the primer are the homology arm of the 23-72 position of the open reading frame of the LacZ gene (underlined), and the latter base is the front part of the sacB gene on the pSNR plasmid ;ZDE2:5'- TGGTAATGGTAGC GACCGGCGCTCAGCTGGAATTCCGCCGATACTGACGG TCAGAAGAACTCGTCAAGAAG-3'(SEQ ID NO.4), the first 50 bases of the primer are the homology arm of the 23-72 position in the opposite direction of the open reading frame of the LacZ gene (indicated by the underline), and the following bases are the neo gene on the pSNR plasmid backend part. A product of 2.7 kb was obtained by PCR amplification, and the PCR amplification system, reaction conditions and recovery method were the same as above.
[0083] 2. The sacB-neoPCR prod...
Embodiment 3
[0107] Example 3. LacZ gene point mutation
[0108] 1. Amplification of the sacB-neo gene cassette with homology arms
[0109] Design primer ZPM1: 5′- ACTGGCCGTCGTTTTACAACGTCGTGACTGGGAA A ACCCTGGCGTTACCC ATCCTTTTATGATTTTCTATC-3' (SEQ ID NO.10), the first 50 bases of the primer are the homology arms of the 20-70 positions of the open reading frame of the LacZ gene (underlined, the latter 22 bases are the front end of the sacB gene on the pSNR plasmid part; ZPM2:5′-G CTATTACGCC AGCTGGCGAAAGGGGGATGTGCTGCAAGGCGATTAAGTT TCAGAAGAACTCGTCAAGAAG-3', (SEQ ID NO.11), the first 50 bases of the primer are the homology arms of the 20-70 position in the opposite direction of the open reading frame of the LacZ gene (indicated by the underline), and the following bases are the neo on the pSNR plasmid The back end of the gene.
[0110] A product of 2.7 kb was obtained by PCR amplification, and the PCR amplification system, reaction conditions and recovery method were the same as above....
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com