Biological sweet protein monellin gene
A technology of sweetness and protein, applied in the direction of plant peptides, plant products, botanical equipment and methods, etc., can solve the problem of no research reports on sweet protein genes, so as to improve the utilization value, nutritional value and eating quality, and improve Effect of protein content and sweetness
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0028] Cloning of the monellin gene:
[0029] (1) According to the amino acid sequence of the monellin protein reported by Kondo K etc. (Kondo, K., Y.Miura, et al. High-level expression of a sweet protein, monellin, in the food yeast Candida utilis.NatBiotechnol, 1997, 15 (5): 453-7), according to the codon preference of Pichia pastoris and Morus alba, a 294bp sweet protein monellin gene was synthesized;
[0030] (2) Artificially synthesize the sequence onto the PUC57-T cloning vector;
[0031] (3) After obtaining positive clones, send them to GenScript for sequencing.
Embodiment 2
[0033] Recombination of monellin gene in Pichia pastoris GS115:
[0034] (1) The T clone containing the target fragment and the pPIC9K empty plasmid were double digested with EcorI and NotI respectively, and the target fragments were recovered respectively, connected with T4 DNA ligase and transformed into E. coli DH5α competent cells, and obtained by screening pPIC9K containing -cloning of monellin, and then send the cultured bacterial liquid to GenScript for sequencing to prove that the monellin gene has been connected to the expression vector pPIC9K;
[0035] (2) Extract the plasmid with the correct sequence for linearization and dephosphorylation, and cultivate fresh yeast GS115 at the same time to prepare competent; then transform the plasmid (pPIC9K expression vector with monellin gene) treated in the previous step into yeast by electric shock transformation method GS115;
[0036] (3) The colonies grown in the MD medium were planted on the YPD plate containing G418 to s...
Embodiment 3
[0039] Blind test of target protein sweetness:
[0040] (1) control sample: mass ratio 10% sucrose aqueous solution
[0041] (2) Preparation of series concentration experimental samples:
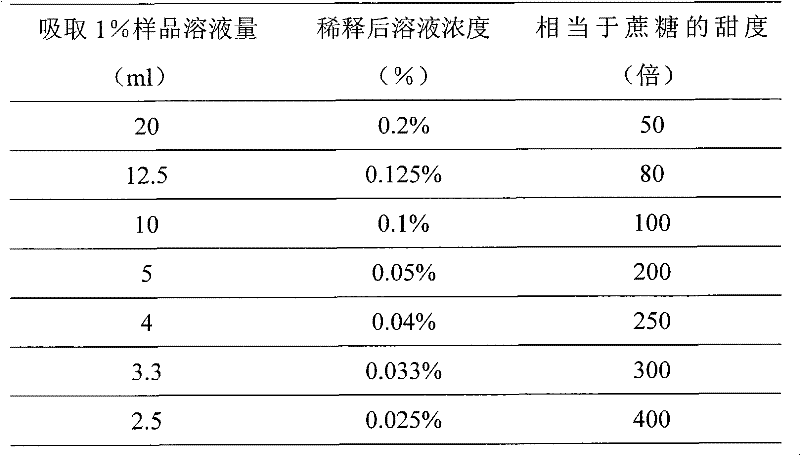
[0042] First weigh 1.0g of the target protein sample and dilute it to 100ml with distilled water to form a 1.0% sample solution; measure 20ml, 12.5ml, 10.0ml, 5ml, 4ml, 3.3ml, 2.5ml of the 1.0% sample solution into a 100ml volumetric flask , diluted to 100ml, the solution concentrations are 0.2%, 0.125%, 0.1%, 0.05%, 0.04%, 0.033%, 0.025% respectively;
[0043](3) Sample tasting comparison: An evaluation team composed of at least 5 people tastes the samples, and judges which of the series concentration samples has the same or similar sweetness as the 10% sucrose solution, so as to calculate the sweetness of the samples. The formula is: Sweetness = 10% / sample concentration. The blind test results of the sweetness of the target protein are as follows:
[0044] Table 1 Relationship between ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com