Method for quantitatively detecting deoxyribonucleic acid (DNA) demethylation capability of pollutant
A quantitative detection method and demethylation technology, applied in the direction of biochemical equipment and methods, microbial determination/inspection, etc., can solve the problem of inability to quantify the demethylation ability of pollutants, quantitative detection, no detection technology, etc. problems, to achieve the effect of easy popularization and application, low requirements for hardware facilities, and simple steps
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
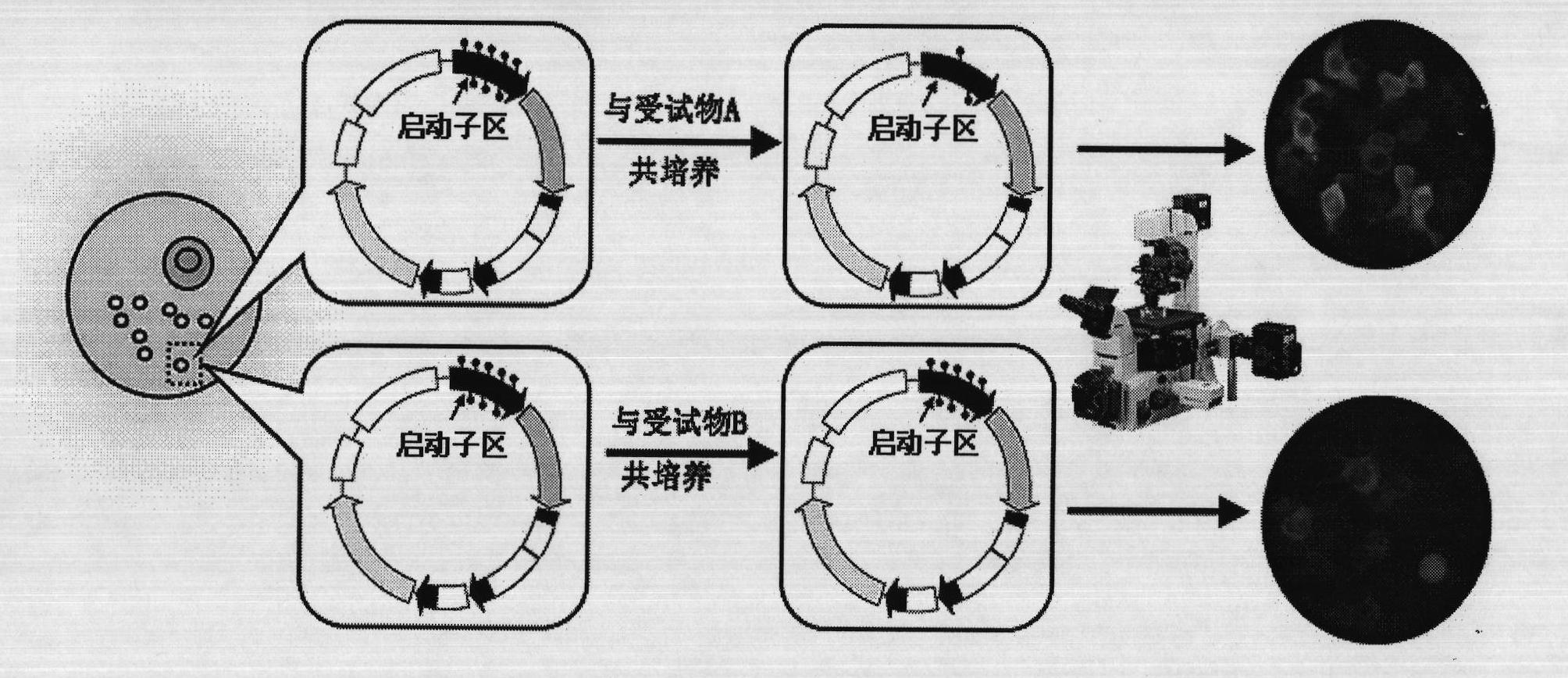
[0030] The present invention will be further described in detail by taking the quantitative detection of the demethylation ability of edible aquatic products polluted by the coast as an example. Specific steps are as follows:
[0031] (1) Methylation treatment of fluorescent plasmid
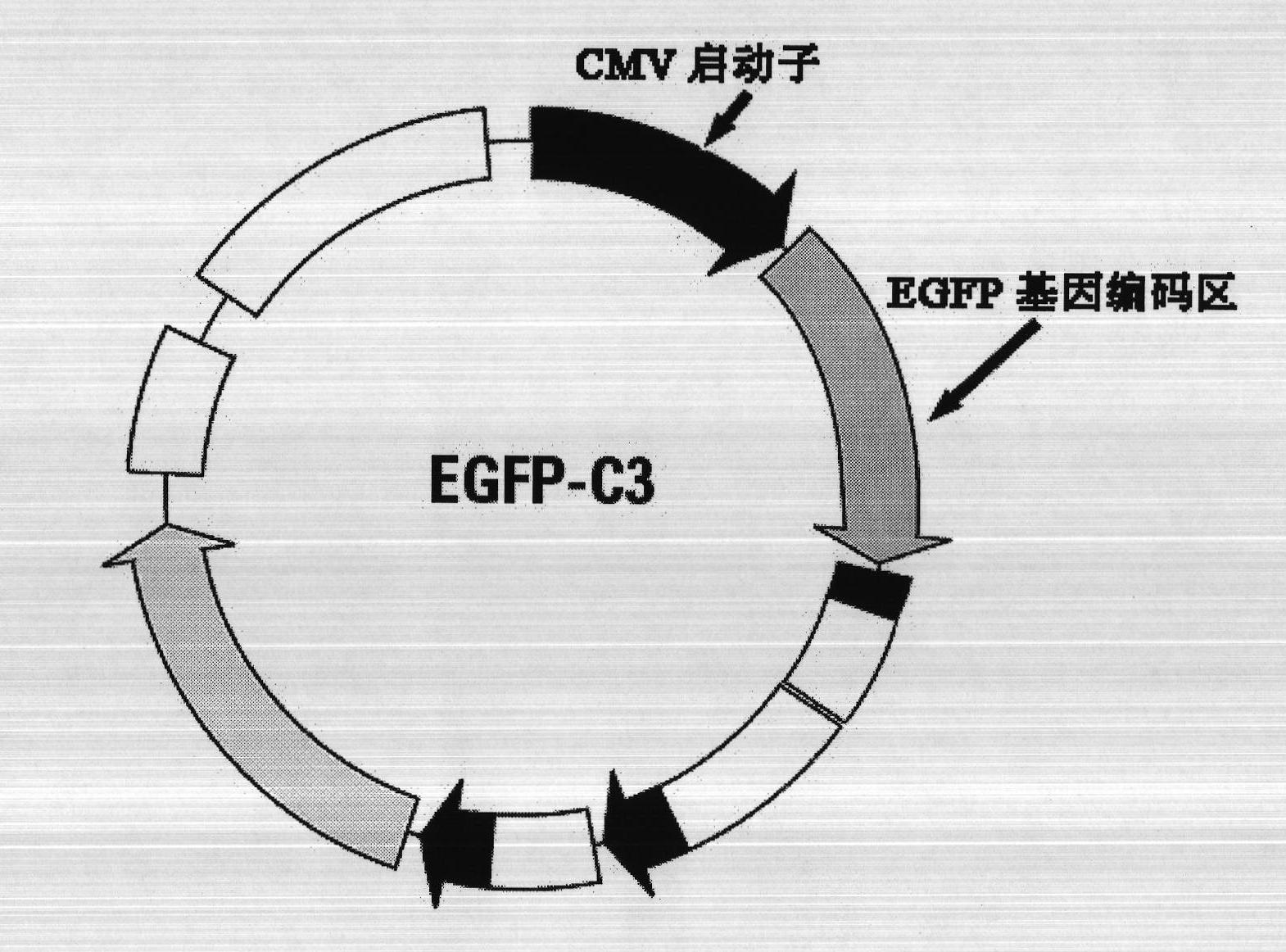
[0032] Purchase the EGFP-C3 plasmid (hereinafter referred to as the C3 plasmid) from Clontech Company of the United States. Purchase DNA methylase Msss.I from NEB Company of the United States. Mix the C3 plasmid with DNA methylase Msss.I, etc., and incubate at 37°C for 3 hours, so that the CMV promoter region of the EGFP gene in the C3 plasmid is in a hypermethylated state. Specific reaction system composition: 4 μl buffer stock solution (10×NE Buffer2), 4 μl S-adenosylmethionine stock solution (100×SAM), 8 μl C3 plasmid DNA (1.7 μM), 6 μl methylase M.SssI, Supplement 18 μl of double distilled water to 40 μl (10×NE Buffer2 buffer stock solution, 100×SAM adenosylmethionine stock solution are pr...
Embodiment 2
[0048] Taking the quantitative detection of demethylation ability of groundwater samples as an example, the present invention will be further described in detail below. Specific steps are as follows:
[0049] (1) Methylation treatment of fluorescent plasmid
[0050] Purchase the EGFP-C3 plasmid (hereinafter referred to as the C3 plasmid) from Clontech Company of the United States. Purchase DNA methylase Msss.I from NEB Company of the United States. Mix the C3 plasmid with DNA methylase Msss.I, etc., and incubate at 37°C for 3 hours, so that the CMV promoter region of the EGFP gene in the C3 plasmid is in a hypermethylated state. Specific reaction system composition: 4 μl buffer stock solution (10×NE Buffer2), 4 μl S-adenosylmethionine stock solution (100×SAM), 8 μl C3 plasmid DNA (1.7 μM), 6 μl methylase M.SssI, Supplement 18 μl of double distilled water to 40 μl (10×NE Buffer2 buffer stock solution, 100×SAM adenosylmethionine stock solution are provided free of charge with...
Embodiment 3
[0066] Taking the quantitative detection of demethylation ability of surface water samples as an example, the present invention will be further described in detail below. Specific steps are as follows:
[0067] (1) Methylation treatment of fluorescent plasmid
[0068] Purchase the EGFP-C3 plasmid (hereinafter referred to as the C3 plasmid) from Clontech Company of the United States. Purchase DNA methylase Msss.I from NEB Company of the United States. Mix the C3 plasmid with DNA methylase Msss.I, etc., and incubate at 37°C for 3 hours, so that the CMV promoter region of the EGFP gene in the C3 plasmid is in a hypermethylated state. Specific reaction system composition: 4 μl buffer stock solution (10×NE Buffer2), 4 μl S-adenosylmethionine stock solution (100×SAM), 8 μl C3 plasmid DNA (1.7 μM), 6 μl methylase M.SssI, Supplement 18 μl of double distilled water to 40 μl (10×NE Buffer2 buffer stock solution, 100×SAM adenosylmethionine stock solution are provided free of charge wi...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com