Virtual screening method for novel cancer-preventing or anti-cancer medicament by taking Keap1 as target point
An anti-cancer drug, virtual screening technology, applied in special data processing applications, instruments, electrical digital data processing, etc., can solve the problems of long cycle, low positive rate, complex natural drugs and food ingredients, etc. The effect of speed and efficiency
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0038] Example 1 A virtual screening of chemical anti-cancer and anti-cancer drugs using the cysteine residue Cys151 of the keap1 protein as the active site.
[0039] (1) Virtual screening steps are as follows:
[0040] 1. Using homology modeling software to determine the structural data of the PDB of the structural activity domain of Keap1 protein
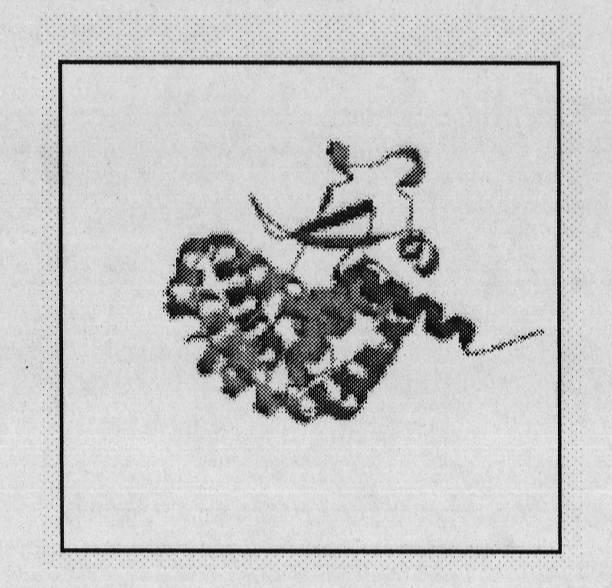
[0041] Using the Swiss Model online service, extract the amino acid sequence of human Keap1 (FASTA format, figure 2 ), submitted the template search function of the Swiss Model, and used the protein tertiary structure of KLHL11 (3i3nb), which is highly conserved with the BTB functional domain of keap1, as a template to perform homology modeling to determine the protein tertiary structure of the active functional domain of keap1 (1 -314) such as image 3 shown.
[0042] The models obtained by the homology modeling method often have some unreasonable structures, so the online model analysis tool Molprobity (http: / / molprobity.b...
Embodiment 2
[0089] Example 2 A virtual screening of chemical anti-cancer and anti-cancer drugs using the cysteine residues Cys273 and Cys288 of the keap1 protein as active sites.
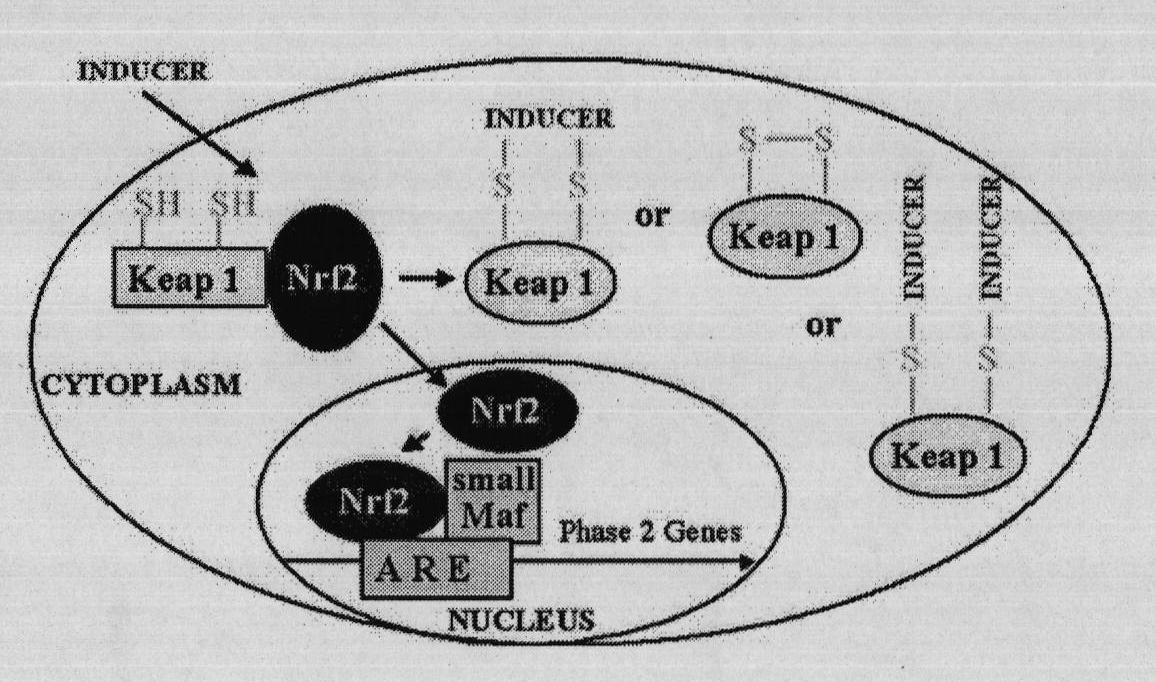
[0090] When the Michael addition reaction of Cys273 and Cys288 of Keap1 protein occurs at the same time, the configuration of Keap1 can be changed, and the Nrf2 transcription factor can be dissociated accordingly, so that it can enter the nucleus to increase the expression of phase II enzymes. According to this basic theory, it is determined that two cysteine residues, Cys273 and Cys288, are simultaneously active centers.
[0091] 1. Using homology modeling software to determine the structural data of the PDB of the structural activity domain of Keap1 protein
[0092] Same as Example 1
[0093] 2. Use Autodock molecular docking software to determine the active center and set the active pocket according to the active cysteine residue site CYS151 of keap 1;
[0094] With embodiment 1, difference is:
[0...
Embodiment 3
[0120] Example 3 A virtual screening of chemical anti-cancer and anti-cancer drugs using the cysteine residue Cys257 of the keap1 protein as the active site.
[0121] The method is the same as that of Example 1, except that: when setting the active pocket: the center coordinate of Cys257 is used as the center of the active pocket.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com