Electrochemical detection method for DNA three-dimensional nanostructure probe
A three-dimensional nano- and nano-structure technology, applied in biochemical equipment and methods, microbial determination/inspection, measuring devices, etc., can solve the problems of strict control of probe distance and unfavorable DNA biological activity, etc., to achieve high selectivity, Effects to avoid interactions and improve performance
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0042] Reagents include:
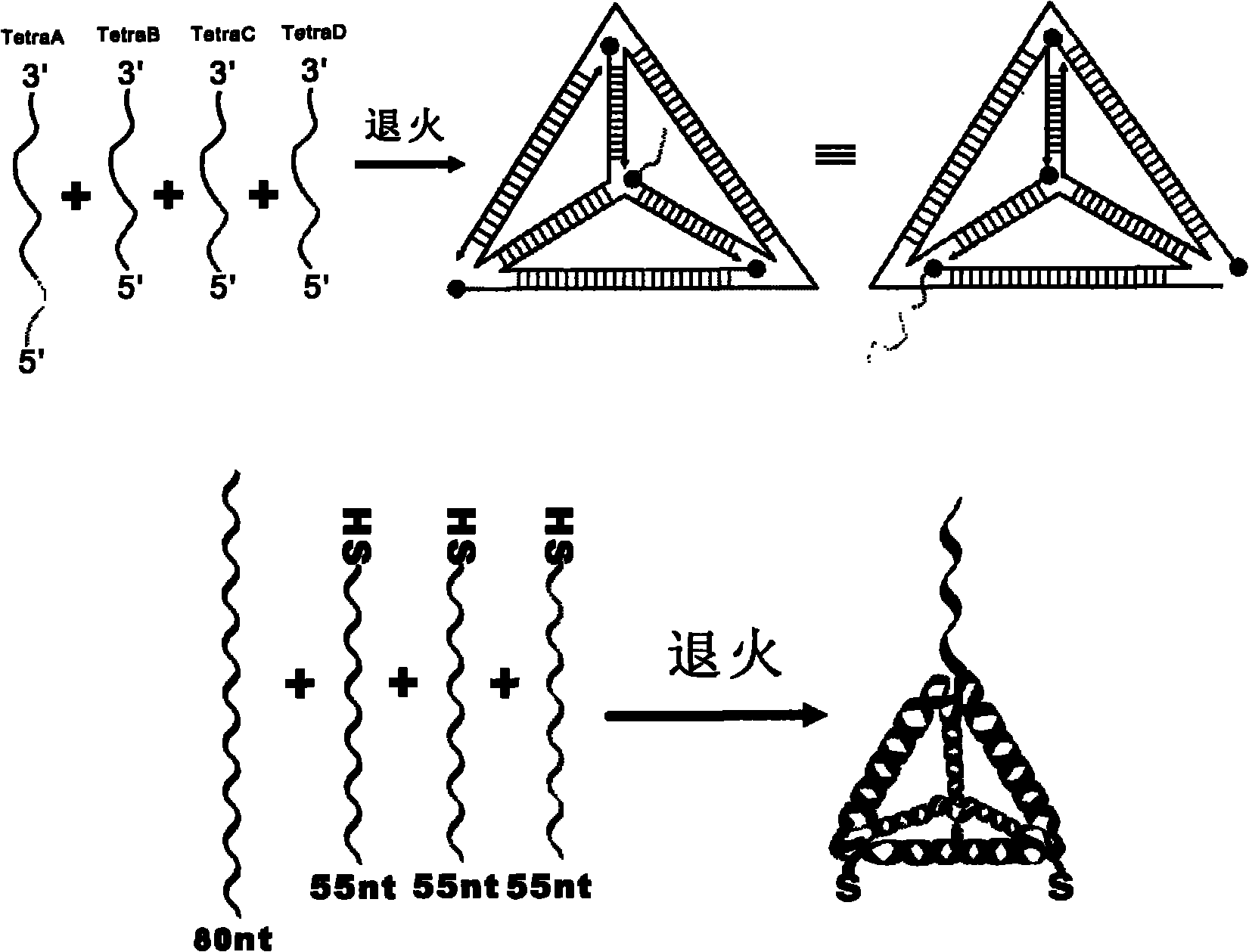
[0043] 4 pieces of single-stranded DNA used to assemble tetrahedral DNA nanostructure probes, Tetra-A (80bp, molecular weight 24539.0, ssDNA), Tetra-B (55bp, molecular weight 17018.0, 5' end modified sulfhydryl ssDNA), Tetra-C (55bp, molecular weight 16898.0, ssDNA modified with thiol at the 5' end), Tetra-D (55bp, molecular weight 16877.0, ssDNA modified with thiol at the 5' end), both were purchased from Dalian Takara Biological Co., Ltd.
[0044] The four DNAs that constitute the tetrahedral structure probe contain three structural domains, each of which is complementary to the corresponding domains of the other three single-stranded DNAs (17 pairs of bases are complementary), and each single-stranded DNA surrounds the tetrahedral structure One side of a circle, each vertex contains two bases (non-complementary, flexible) for bending, the 3' end and 5' end of the single-stranded DNA converge at the four vertices of the tetrahedron, and TetraA is a...
Embodiment 2
[0080] Reagents include:
[0081] DNA is the same as Example 1
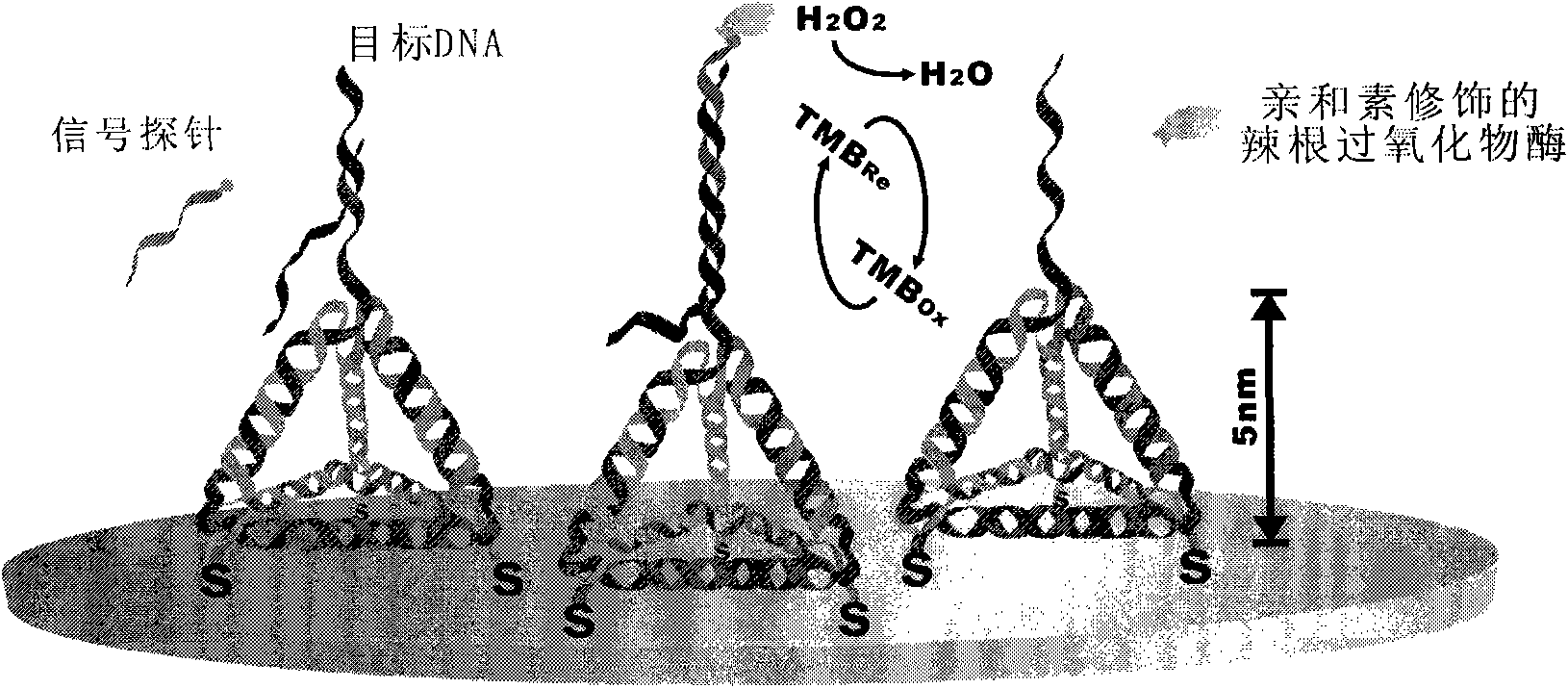
[0082] Avidin-modified glucose oxidase (GOx-A), purchased from Vector Laboratories (SanDiego, CA)
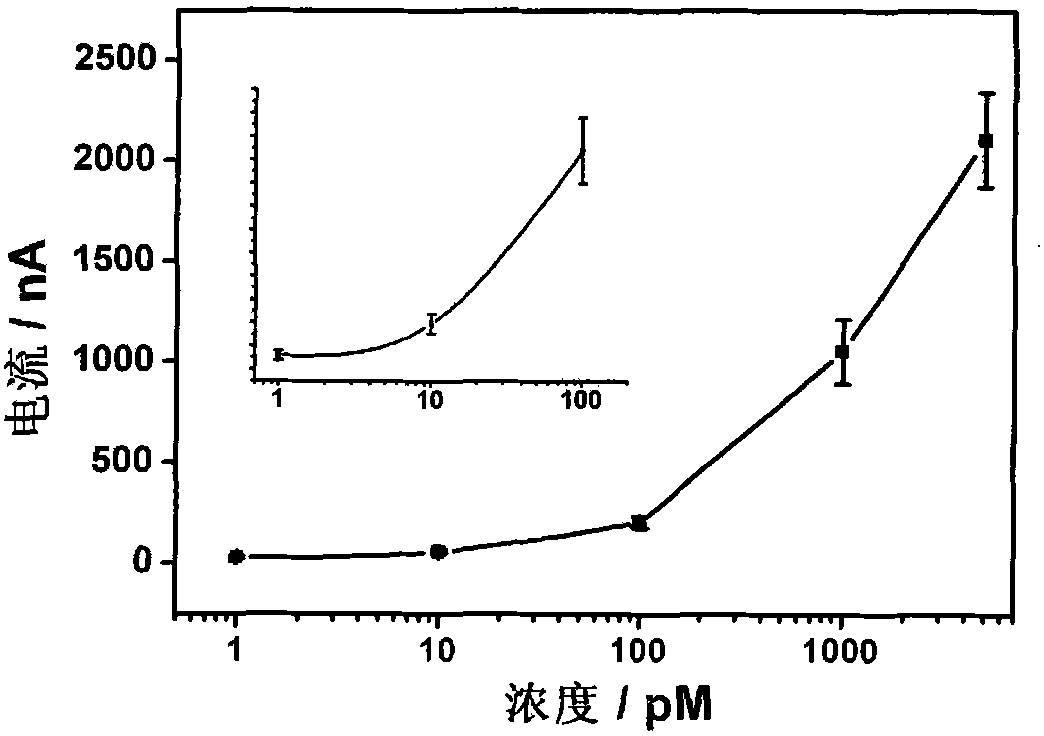
[0083] The experimental procedure is the same as that in Example 1, except that the avidin-modified horseradish peroxidase is replaced by avidin-modified glucose oxidase, and the substrate TMB is replaced by glucose and benzoquinone. Voltage is 0.35V (reference is silver / silver chloride electrode), all the other parameters are constant, the result is the same as that of Example 1.
Embodiment 3
[0085] Reagents include:
[0086]4 pieces of single-stranded DNA used to assemble tetrahedral DNA nanostructure probes, S1 (88bp, molecular weight 27014.0, ssDNA), S2 (83bp, molecular weight 25642.6, ssDNA modified with thiol at the 5' end), S3 (83bp, molecular weight 25678.6, 5' end modified thiol ssDNA), S4 (83bp, molecular weight 25535.5, 5' end modified thiol ssDNA), were purchased from Dalian Takara Biological Co., Ltd. S1:
[0087] 5'-GTATCCAGTGGCTCATTTTTTTTTACGAACATTCCTAAGTCT
[0088] GAAATTTATCACCCGCCATAGTAGACGTATCACCAGGCAGTTG
[0089] AG-3'
[0090] S2:
[0091] 5’-HS-ATTCAGACTTAGGAATGTTCGACATGCGAGGAGGAAATG
[0092] AAGTCCAATACCGACGATTACAGGCCTTTGCGCCTTGCTACAC
[0093] G-3'
[0094] S3:
[0095] 5’-HS-ACGTGTAGCAAGGCGCAAAGGCCTGTAATCGACTCTAC
[0096] GGGAAGAGCATGCCCATCCGGCTCACTACTATGGCGGGTGAT
[0097] AAA-3'
[0098] S4:
[0099] 5’-HS-ACTCAACTGCCTGGTGATACGAGAGCCGGATGGGCATG
[0100] CTCTTCCCGTAGAGACGGTATTGGACTTCATTTTCCTCCTCGCA
[0101] TG-3'
[0102] in,
...
PUM
| Property | Measurement | Unit |
|---|---|---|
| diameter | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com