Method for designing oligonucleotide chip probe and HIV oligonucleotide probe and quality control method thereof
A technology of oligonucleotide probes and design methods, applied in the field of detection reagents, can solve problems such as the inability to directly and effectively evaluate chip quality, and achieve high sensitivity and high efficiency
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
example 1
[0026] This example adopts the oligonucleotide probe design method of the present invention to prepare a kind of human immunodeficiency virus (HIV-1) oligonucleotide probe, and its preparation method and process are as follows:
[0027] 1. First, obtain the full-length cDNA sequence of human immunodeficiency virus (HIV-1) B subtype U26942 from the GenBank database, and obtain the conserved sequence of HIV virus through BLAST analysis, and use the biological software Oligo 6.0 to retrieve the obtained BLAST sequences respectively. The conserved sequence of HIV-1B subtype U26942 was analyzed one by one, and an Oligo probe with a length of 50mer was designed according to the design principles of oligonucleotide probes, and its nucleotide sequence was
[0028] ATAGTATGGGCAAGCAGGGAGCTAGAACGATTCGCAGTTAATCCTGGCCT;
[0029] 2. Then, use the biological software Oligo 6.0 to design a universal sequence with a length of 10mers according to the design principles of oligonucleotide probes:...
example 2
[0042] This example is an example of the application of the human immunodeficiency virus oligonucleotide probe obtained in Example 1 in the quality control of the human immunodeficiency virus chip.
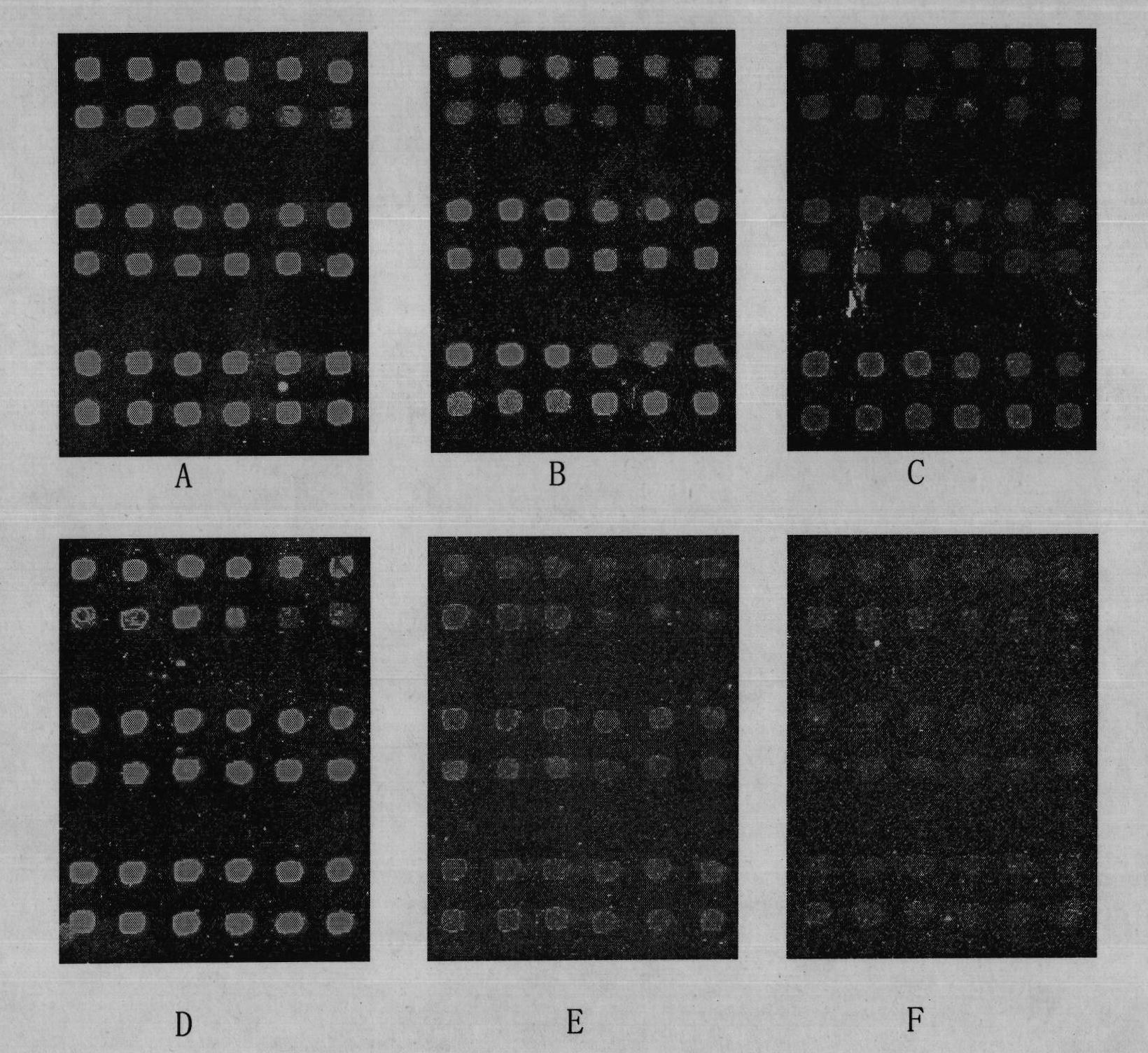
[0043] First, the human immunodeficiency virus oligonucleotide probe obtained in Example 1 was synthesized by a DNA synthesizer, dissolved in the spotting solution, adjusted to a final concentration of 1 μg / μl, and transferred to a 384-well plate for use. Using the Cartesian PixSys 5500 gene chip spotting instrument, the above probes and blank controls were printed on the epoxy-coated glass slides of Corning Company, in which 6 probe points were printed in each line, and the blank controls were printed in lines 3 and 6. To prepare and obtain biochips, the above-mentioned spotting solution is Pronto! TM For epoxy spotting solution, the spotting solution is used as the blank control solution, and the signal of the blank control can represent the spotting process of the chip and the...
PUM
| Property | Measurement | Unit |
|---|---|---|
| melting point | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com