Gastric cancer detection marker and detecting method thereof, kit and biochip
A technology for detecting markers and biochips, which can be used in biochemical equipment and methods, microbial determination/inspection, DNA/RNA fragments, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
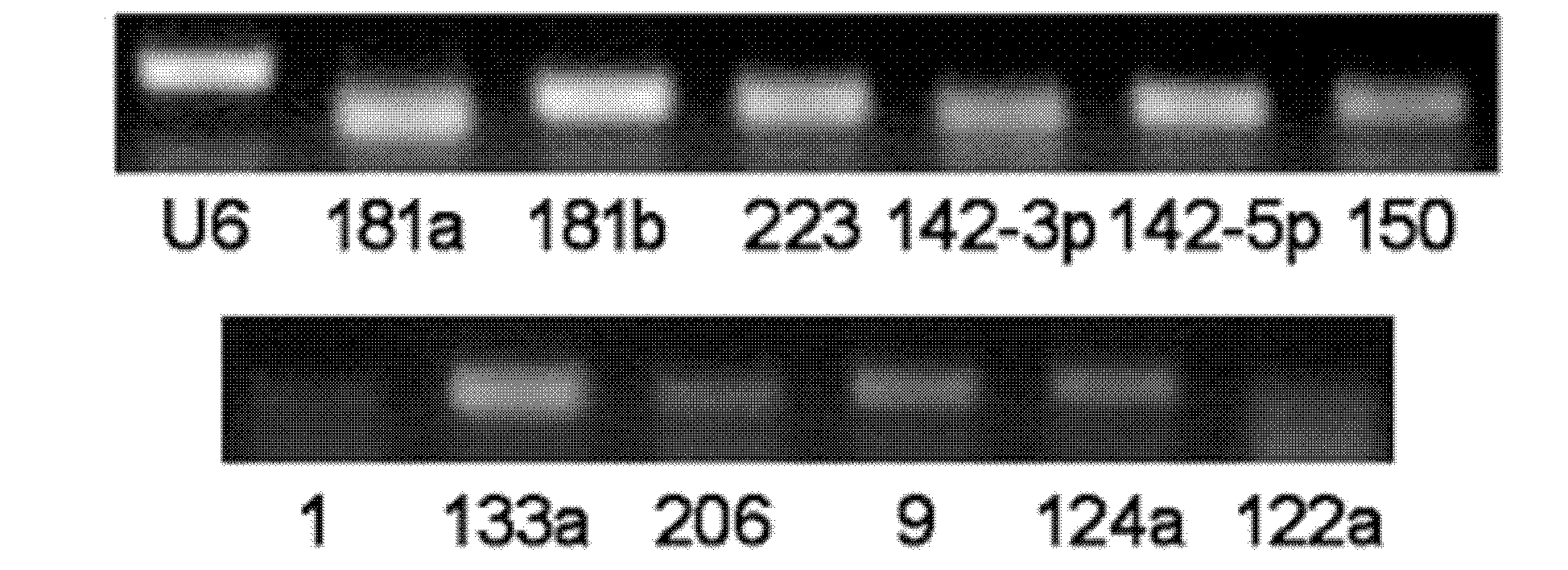
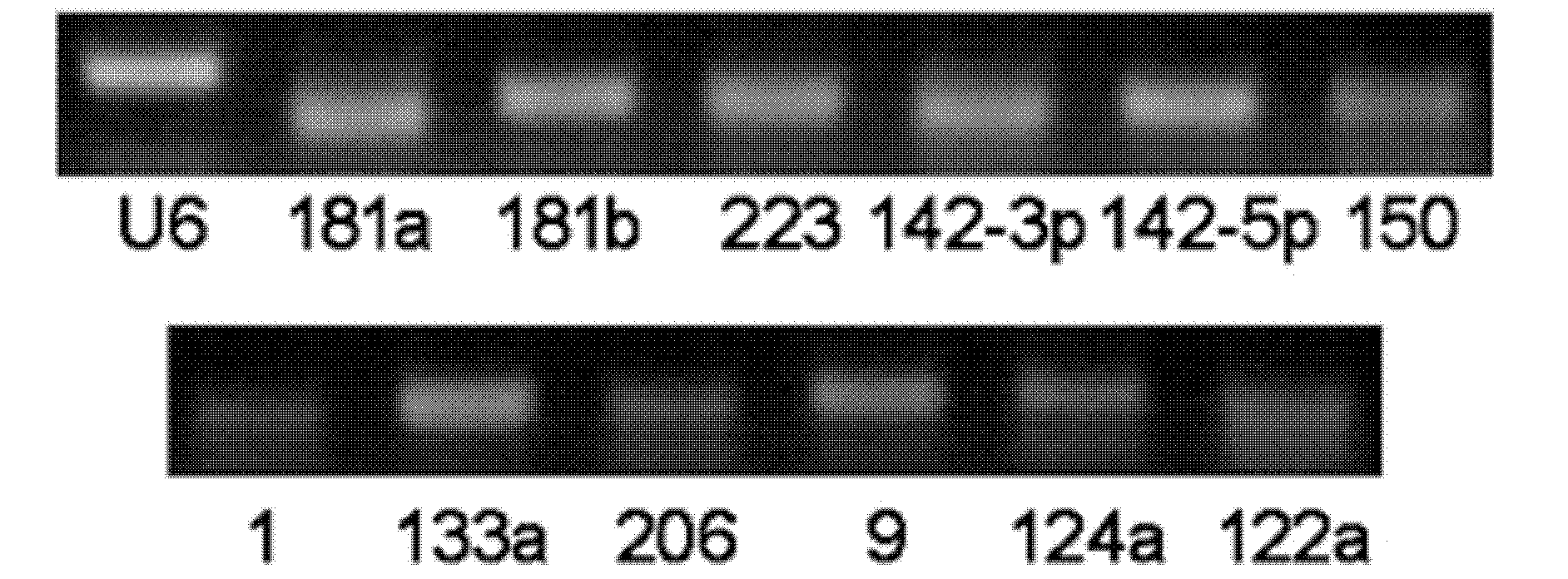
[0083] The RT-PCR experiment of microRNA in embodiment 1 serum / plasma
[0084] Using RT-PCR technology to discover and prove that various microRNAs exist stably in human and animal serum / plasma, and their expression is quite abundant. The specific steps are:
[0085] (1) Collect serum / plasma from mice, rats, normal people and certain patients;
[0086] (2) Preparation of cDNA samples. There are two schemes for this operation, one scheme is to directly carry out reverse transcription reaction on 10 μl serum / plasma, and the other is to use Trizol reagent (Invitrogen Company) to first extract serum / plasma total RNA (10ml serum / plasma can usually enrich about 10 μg Left and right RNA), and then cDNA was obtained by RNA reverse transcription reaction. The reverse transcription reaction system included 4 μl 5×AMV buffer, 2 μl 10 mMeach dNTP mixture (Takara), 0.5 μl RNase Inhibitor (Takara), 2 μl AMV (Takara) and 1.5 μl gene-specific reverse primer mixture. The reaction steps are...
Embodiment 2
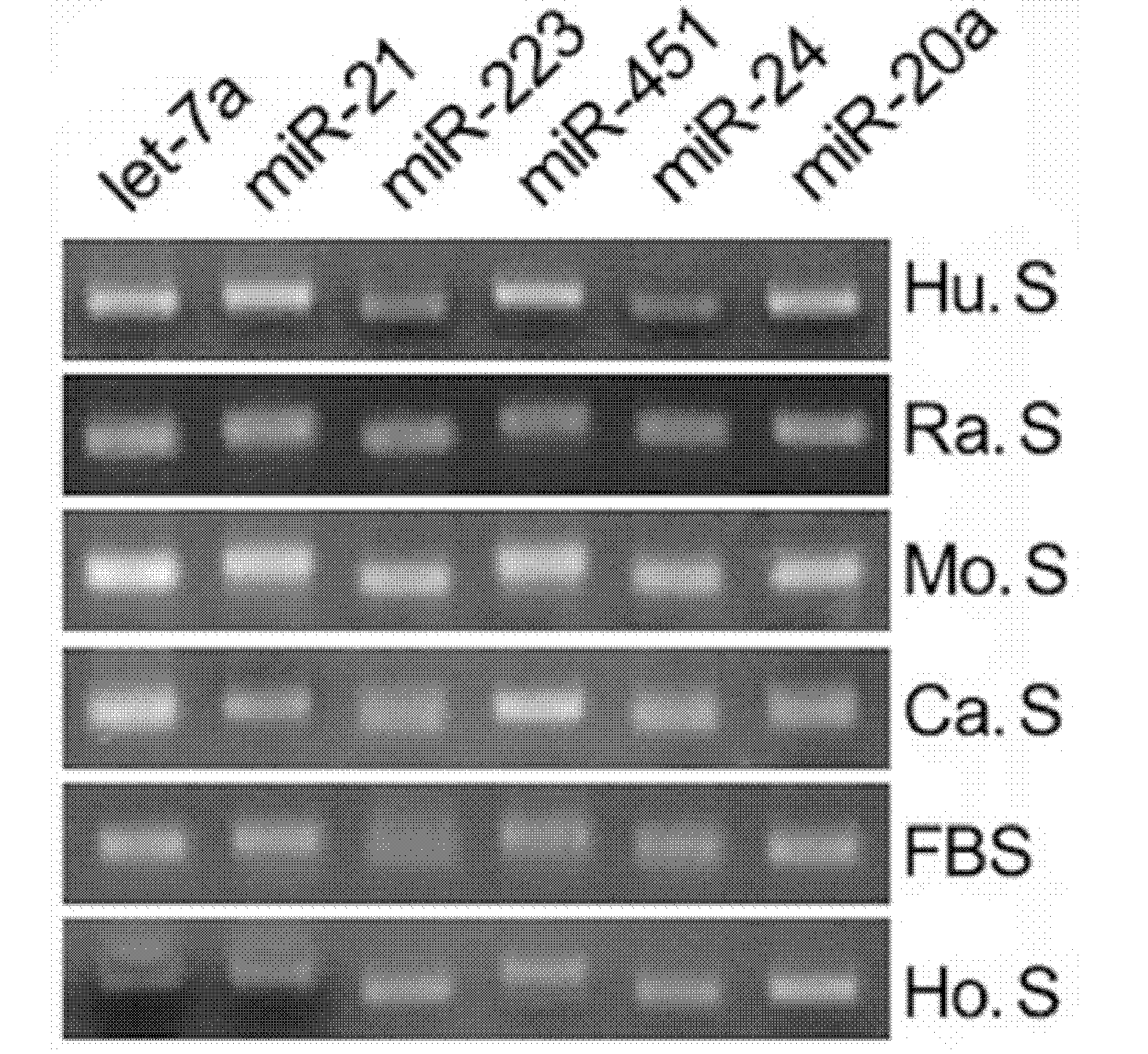
[0092] The real-time PCR experiment of microRNA in embodiment 2 serum / plasma
[0093] In order to study the specific changes of serum / plasma microRNA in the disease process of gastric cancer, the quantitative PCR experiment of serum / plasma microRNA was carried out. The experimental principle and experimental steps of quantitative PCR are the same as RT-PCR, the only difference is that the fluorescent dye EVA GREEN is added during PCR. The instrument used was ABI Prism7300 fluorescent quantitative PCR instrument, and the reaction conditions were 95°C, 5 minutes for 1 cycle → 95°C, 15 seconds, 60°C, 1 minute for 40 cycles. The data processing method is the ΔΔCT method, and CT is set as the cycle number when the reaction reaches the threshold value, then the expression level of each microRNA relative to the standard internal reference can be expressed by equation 2-ΔCT, where ΔCT=CT sample-CT internal reference. The patient's serum / plasma sample and normal human serum / plasma sam...
Embodiment 3
[0095] Embodiment 3 is used for diagnosing the serum / plasma microRNA chip of gastric cancer
[0096] The chip operation process is as follows:
[0097] (1) extract total RNA in serum / plasma, and formaldehyde denaturing gel electrophoresis to detect the quality of total RNA;
[0098] (2) Isolation of microRNA: Take 50-100 μg of total RNA and use Ambion's miRNA Isolation Kit (Cat#.1560) to isolate microRNA;
[0099] (3) Fluorescence labeling of microRNA samples: use T4 RNA ligase labeling method for fluorescent labeling, and then precipitate with absolute ethanol, dry and use for chip hybridization;
[0100] (4) Hybridization and washing: Dissolve RNA in 16 μL of hybridization solution (15% formamide; 0.2% SDS; 3×SSC; 50×Denhardt’s solution), and hybridize overnight at 42°C. After the hybridization, wash in a liquid containing 0.2% SDS and 2×SSC at about 42°C for 4 minutes, then wash in a 0.2×SSC liquid for 4 minutes at room temperature, and the slides can be used for scanning...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com