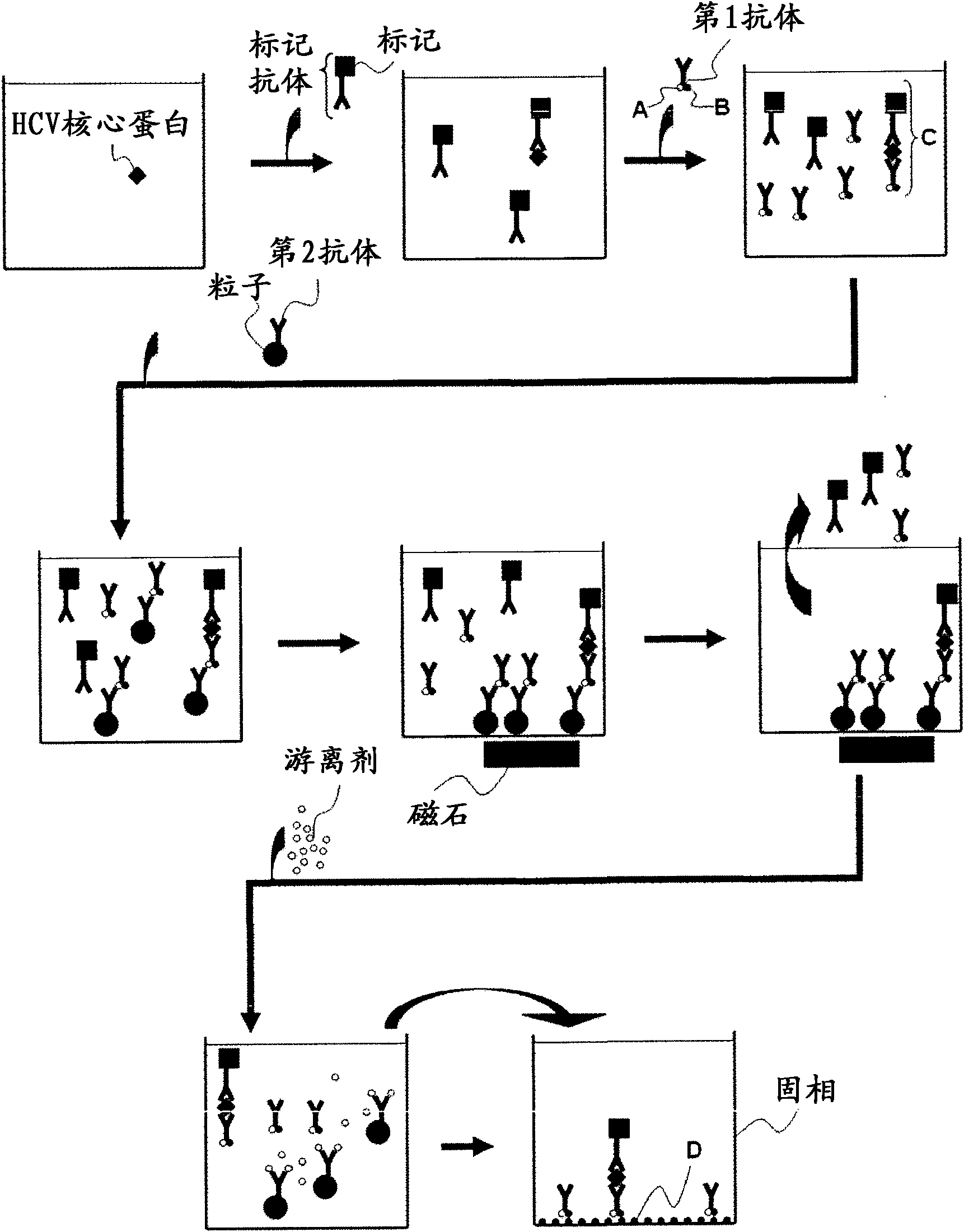
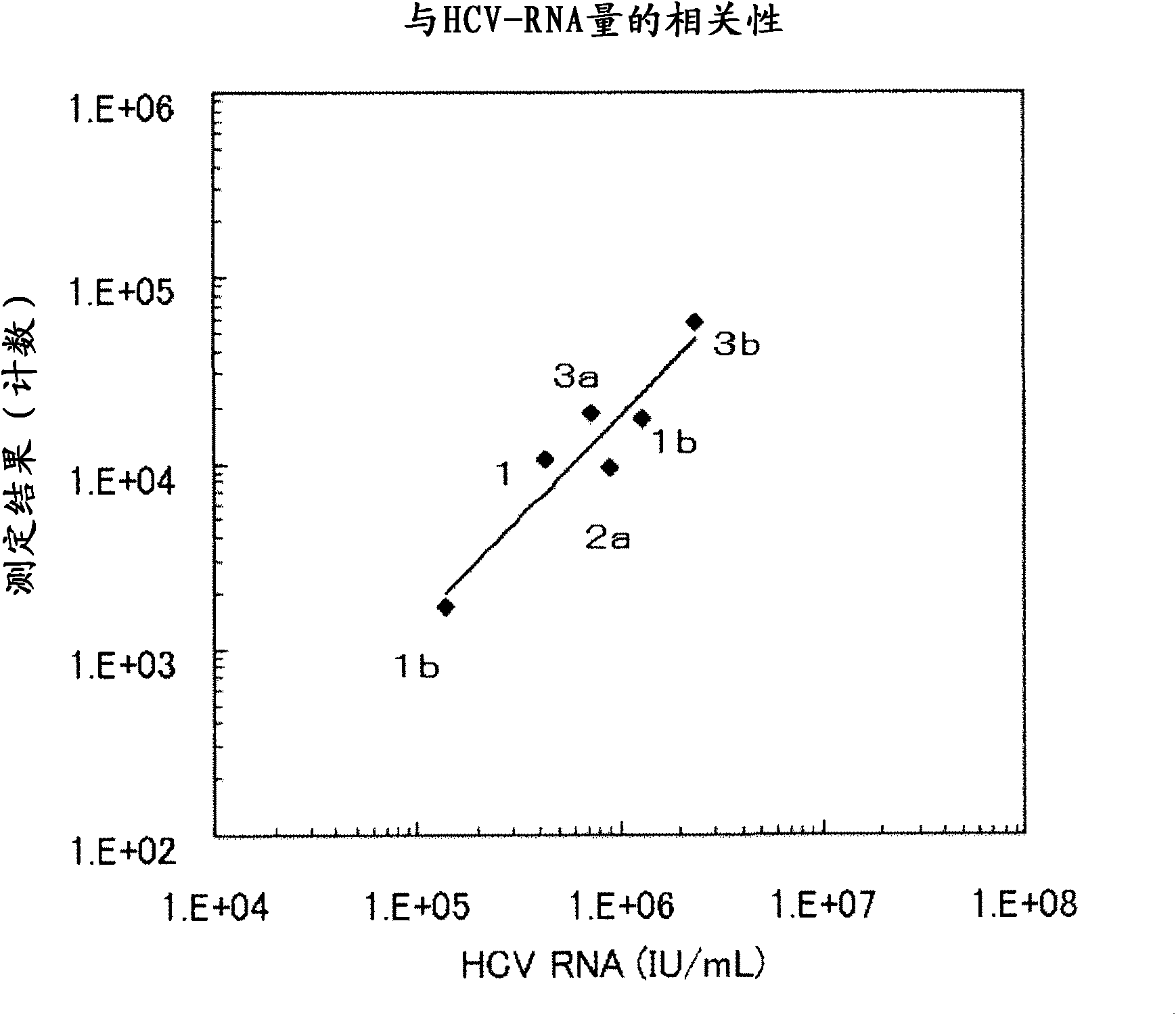
Method for pretreating sample and method for immunoassay of hcv
An immunoassay and sample technology, applied in the field of HCV immunoassay, can solve the problems of high sensitivity, variation, detection, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment approach
[0062] Preferred embodiments of the present invention will be described below with reference to the accompanying drawings.
[0063] The pretreatment method of samples for HCV core protein detection in this embodiment is a pretreatment method for samples used in the detection of HCV core protein using the immunoassay method using particles, including: using samples suspected of containing hepatitis C virus with The first treatment step of the reagent treatment of alkaline substances, the sample obtained in the first treatment step is treated with the reagent containing acidic substances, the second treatment step, at least one of the reagents used in the first treatment step and the second treatment step Contains a reducing agent.
[0064] In this embodiment, at least one of the reagents used in the first treatment step and the second treatment step contains a reducing agent. When at least one of the reagents used in the first treatment step and the second treatment step conta...
Embodiment 1
[0176]
[0177] (A) Construction of expression plasmid
[0178] The following plasmids were obtained by merging the DNA encoding the amino acid sequence at positions 1 to 160 of the HCV core region represented by SEQ ID NO: 2, 3, or 4.
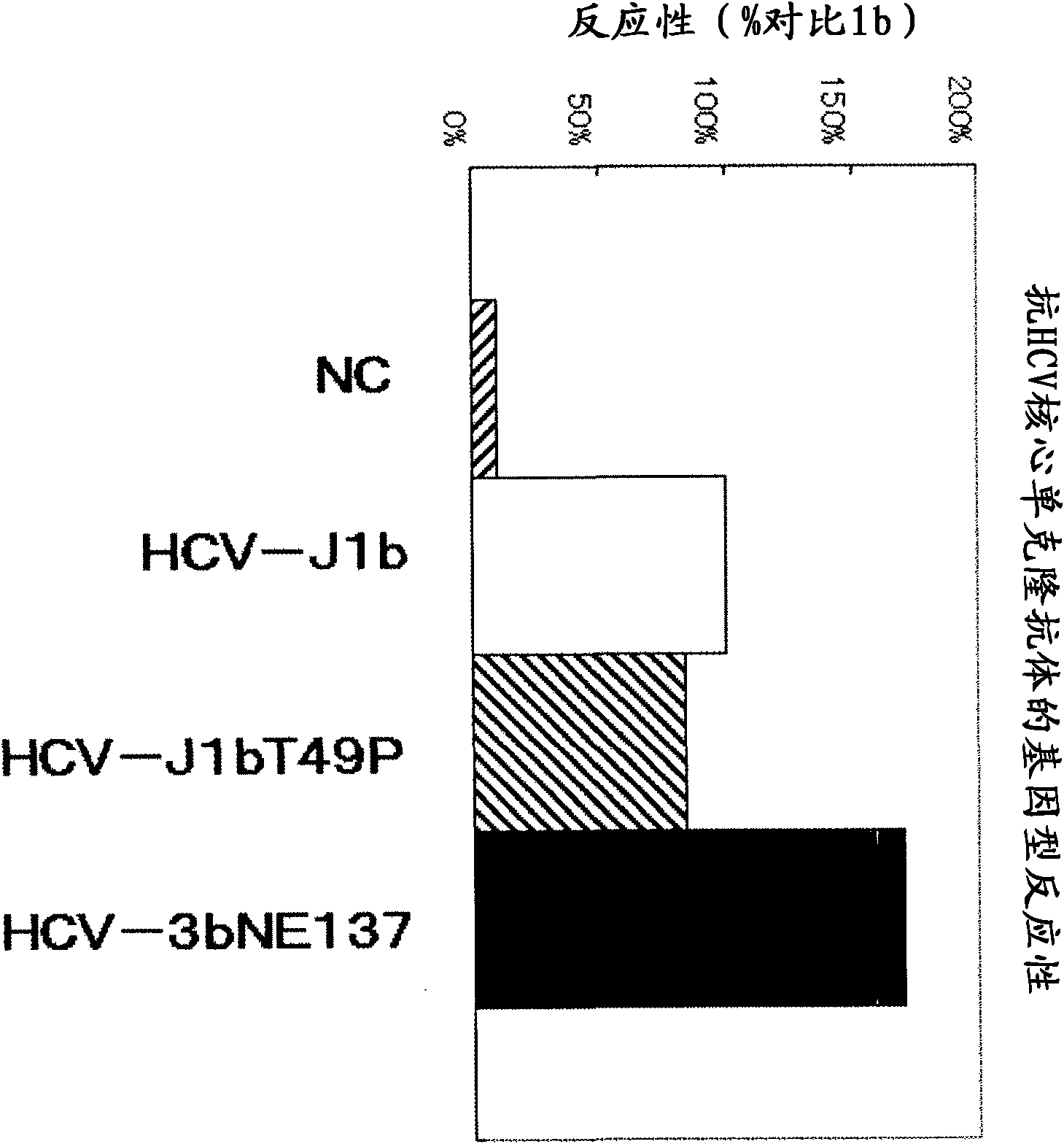
[0179] pUC·HCV-J1b: plasmid of the core region protein (HCV-J1b) of HCV genotype 1b shown in SEQ ID NO: 2
[0180] pUC·HCV-J1bT49P: The plasmid of the core region protein (HCV-J1bT49P) of HCV genotype 1b in which the 49th amino acid threonine shown in SEQ ID NO: 3 is mutated to proline
[0181] pUC·HCV-3bNE137: plasmid of core region protein (HCV-3bNE137) of HCV genotype 3bNE137 strain shown in SEQ ID NO: 4
[0182] SEQ ID NO: 2
[0183] MSTNPKPQRKTKRNTNRRPQDVKFPGGGQIVGGVYLLPRRGPRLGVRATRKTSERSQPRGRRQPIPKARRPEGRTWAQPGYPWPLYGNEGMGWAGWLLSPRGSRPSWGPTDPRRRRSRNLGKVIDTLTCGFADLMGYIPLVGAPLGGAARALAHGVRVLED
[0184] SEQ ID NO: 3
[0185] MSTNPKPQRKTKRNTNRRPQDVKFPGGGQIVGGVYLLPRRGPRLGVRAPRKTSERSQPRGRRQPIPKARRPEGRTWAQPGYPWPLYGNEGMGWAGWLLSPRGSRPSWGPTDP...
Embodiment 2
[0206]
[0207] Using the HCV-J1b and HCV-J1bT49P obtained in Example 1 and the synthetic peptide CDP-2 shown in Table 1, CDP-2-1, CDP-3, and CDP-3-1 were analyzed as anti-HCV core protein single Cloning the epitope of the antibody HCF4-801 antibody and HE25 antibody. It should be noted that CDP-2, CDP-2-1, CDP-3, and CDP-3-1 are synthetic peptides conjugated with keyhole limpet hemocyanin, respectively, which were produced by Operon Biotechnology Co., Ltd. externally.
[0208] Table 1: List of amino acid sequences of peptides derived from HCV core
[0209]
Peptide name
amino acid sequence
AA configuration
SEQ ID NO: 5
CDP-2
DVKFPGGGQIVGGVYLLPRR
21~40
SEQ ID NO: 6
CDP-2-1
DVKFPGGGQI
21~30
SEQ ID NO: 7
CDP-3
GPRLGVRATRKTSKRSQPRG
41~60
SEQ ID NO: 8
CDP-3-1
GPRLGVRATR
41~50
[0210] (A) Confirmation of reactivity between CDP-2 and HCF4-801 antibody
[0211] To contain ...
PUM
| Property | Measurement | Unit |
|---|---|---|
| diameter | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com