Molecular marker identification method for bacterial angular leaf spot resistant gene of cucumber
A technology of molecular markers and identification methods, which is applied in the field of plant biology, can solve the problems of accelerating the process of cucumber breeding and unclear molecular mechanisms, and achieve the effects of improving breeding efficiency, being easy to master, and reducing production costs
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0019] The molecular marker identification method of cucumber anti-bacterial angular leaf spot gene, the method comprises the following steps:
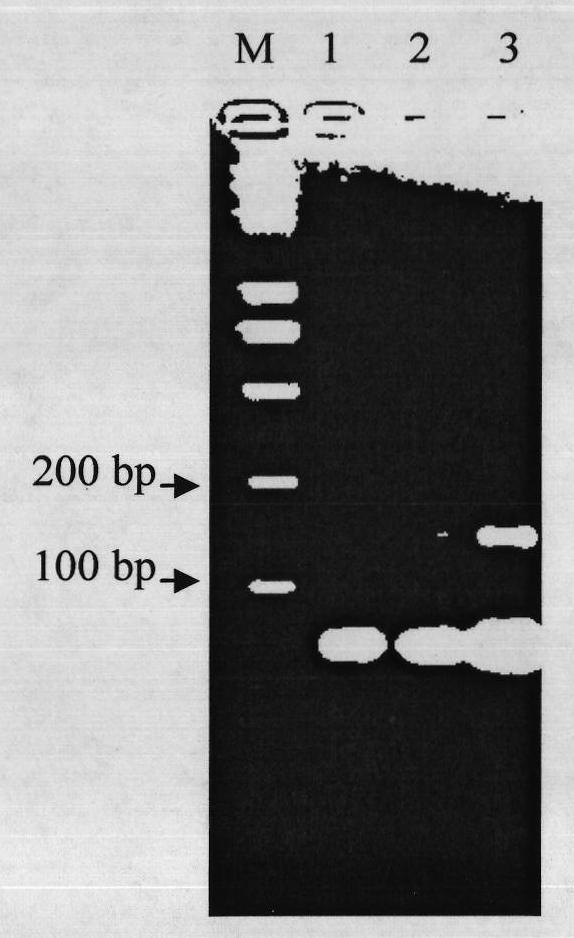
[0020] Use the DNA of a cucumber variety or strain with unknown resistance to bacterial angular spot disease as a template, and use CSWCT24A as a primer to perform PCR amplification, and perform 3% agarose gel electrophoresis on the amplified product or 7.5% agarose gel electrophoresis on the amplified product. Denatured polyacrylamide gel electrophoresis test, if the amplified band type is consistent with the known resistance to bacterial angular spot of cucumber Dongnong 803, then the cucumber variety or line with unknown resistance to bacterial angular spot For a variety or strain resistant to bacterial angular leaf spot, the PCR amplification program is: 94°C pre-denaturation for 5 minutes, 94°C denaturation for 30 seconds, 57°C annealing for 1 minute, 35 cycles, 72°C extension for 1 minute, 72°C extension for 10 minutes, Finally ...
Embodiment 2
[0024] In the molecular marker identification method of the cucumber anti-bacterial angular leaf spot gene, in the PCR amplification, the PCR reaction system of every 20 μl is as follows: 10×buffer: 2 μl, 2mM dNTP: 2 μl, Primer 1 of 10 pmol / μl: 0.8 μl, 10 pmol / μl Primer 2: 0.8 μl, 25 mM MgCl 2 : 2.5 μl, 5 U / μl TaqDNA polymerase: 0.2 μl, 60 ng / μl template DNA: 1 μl, ddH2O: 10.7 μl.
Embodiment 3
[0026] Application of molecular marker identification method of cucumber bacterial angular leaf spot resistance gene in assisted selection of disease-resistant cucumber materials.
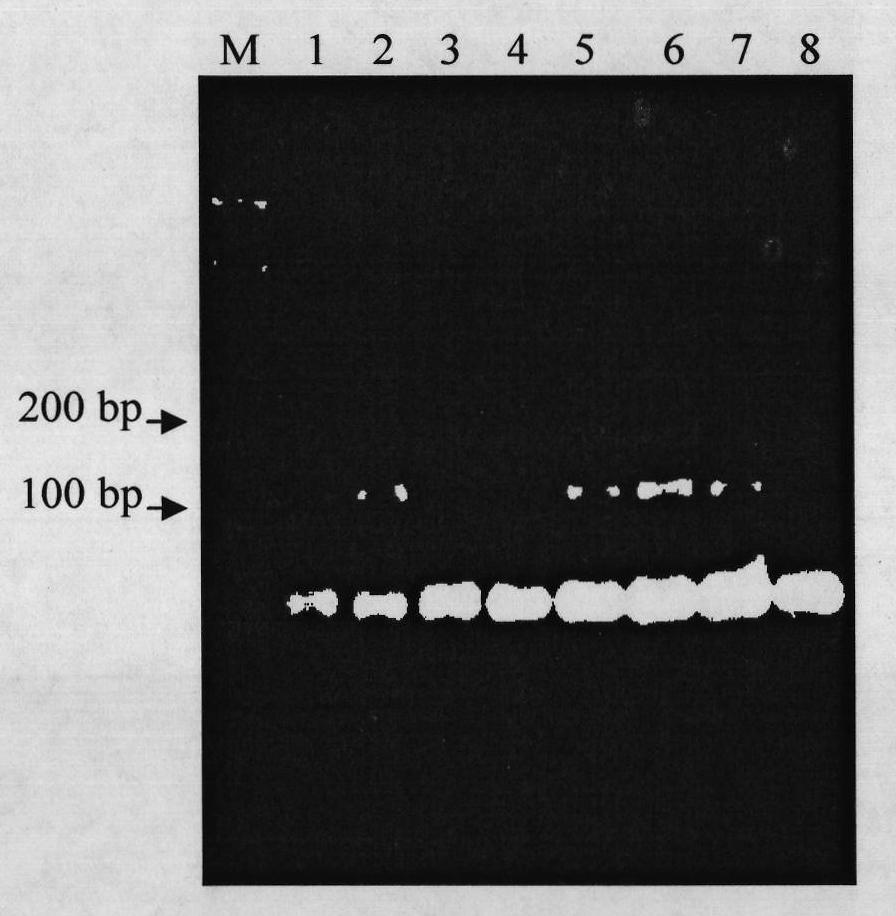
[0027] Using the new cucumber hybrid varieties Dongnong 804 and Dongnong 805 bred by the cucumber research group of the College of Horticulture of Northeast Agricultural University as materials; extracting the DNA of a single plant as a template, and using a specific molecule of cucumber anti-bacterial angular leaf spot provided by the present invention The marker CSWCT24A was used as a primer for PCR amplification. The results showed that the PCR amplification band patterns of Dongnong 804 and Dongnong 805 were completely consistent with the amplified band pattern of the disease-resistant control Dongnong 803, and the results of PCR detection were consistent with those of the field bacteria. The concordance rate of leaf spot resistance identification was 100%, which proved that CSWCT24A is a practi...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com