Application of 3'UTR of ebf1 gene mRNA in repressing gene expression
A technology for suppressing genes and genes, applied in the field of genetic engineering, to achieve the effect of easy cloning
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0018] Example 1 Study on the function of EBF1 mRNA 3'UTR
[0019] Construct a new recombinant gene. According to the 5'→3' direction, a sequence of 643 nt was selected from the stop codon TGA of EBF1, and the target fragment was amplified using Arabidopsis genomic DNA as a template. The nucleotide sequence is shown in SEQ ID No:1. The PCR reaction system used in the amplification process is:
[0020]
[0021] The primer sequences used in the above system are as follows:
[0022] Forward primer 5’-ATCGAATTCTGATCAACAATTCCACTGTC-3’;
[0023] Reverse primer 5'-TGTGGATCCCATGAATAGTCTTAAAGGTG-3'.
[0024] The PCR reaction conditions are:
[0025]
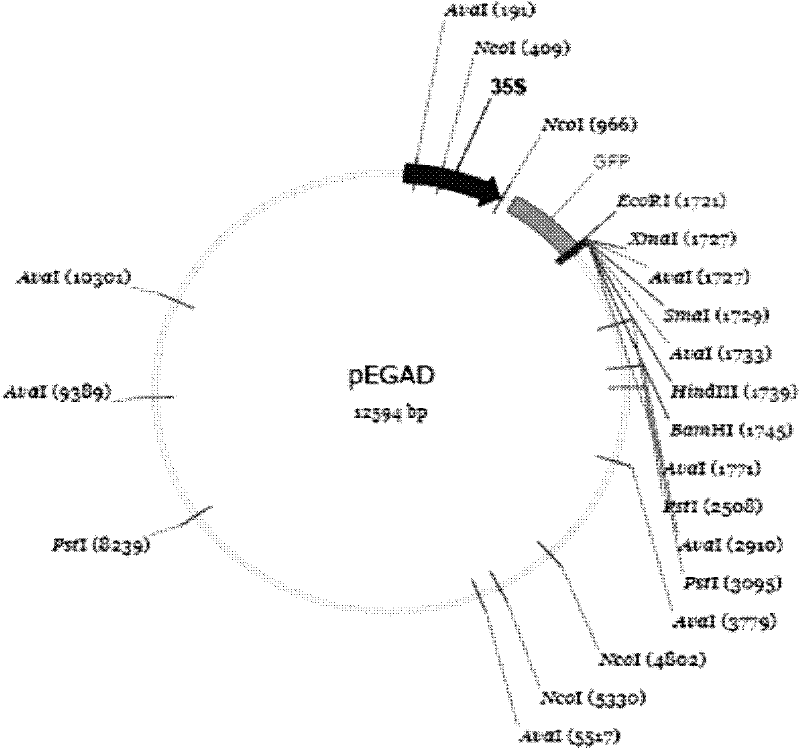
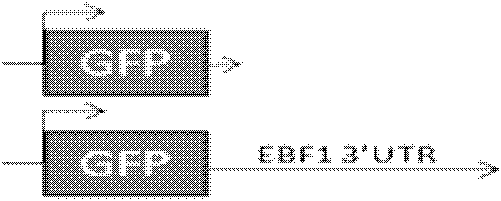
[0026] Then use EcoR I and BamH I two endonucleases to digest the target fragment and the vector pEGAD (such as figure 1 Shown), and then use T4 ligase to insert the target fragment into the vector. The final recombinant gene structure is as follows figure 2 Shown.
[0027] Use Agrobacterium to infect Arabidopsis thaliana inflorescences and transf...
Embodiment 2
[0031] Example 2 Triple reaction of transgenic plants
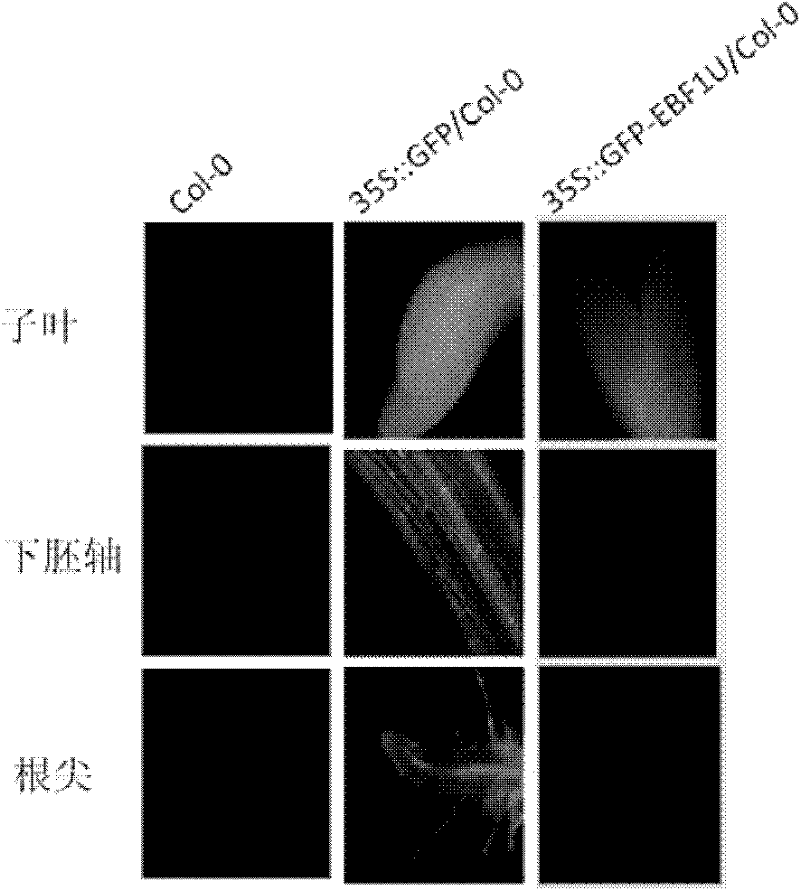
[0032] The classic ethylene reaction is the well-known triple reaction. When ethylene is applied, the chlorotic seedlings show that the hypocotyl becomes shorter and thicker, the root becomes shorter, and the apical hook becomes more intense. The chlorophyll seedlings grown for 3 days on the medium containing 10μM ACC (a precursor of ethylene biosynthesis), 35S::GFP-EBF1U / Col-0 transgenic plants and 35S::GFP / Col-0 transgenic plants Than has obvious ethylene insensitive phenotype, such as Figure 5 Shown. Specific analysis revealed that the latter's triple response phenotype was the same as that of wild-type plants, while the 35S::GFP-EBF1U / Col-0 transgenic plants had significantly longer hypocotyl lengths and root lengths than wild-type plants, and the tip hooks disappeared. This indicates that the 35S::GFP-EBF1U / Col-0 transgenic plant becomes ethylene insensitive due to the overexpression of EBF1 3'UTR.
Embodiment 3
[0033] Example 3 Detection of EIN3 protein level in transgenic plants
[0034] Since the response caused by ethylene signal is activated by the two most critical transcription factors EIN3 / EIL1, the present invention further detects the protein level of transcription factor EIN3 / EIL1 in 35S::GFP-EBF1U / Col-0 transgenic plants. The study found that compared with wild-type 35S::GFP-EBF1U / Col-0 transgenic plants, the protein level of EIN3 / EIL1 was significantly down-regulated, such as Image 6 Shown. The reason why EBF1 3'UTR overexpression becomes ethylene insensitive is because the EIN3 protein level is down-regulated.
[0035] From the above results, it can be seen that if the EBF1 3'UTR is overexpressed in plants, the plants will be insensitive to ethylene, and consistent with this, the level of endogenous EIN3 protein in plants will be down-regulated.
[0036] This gives a good hint that overexpression of EBF1 3'UTR in plants can down-regulate the EIN3 protein level and cause the ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com