Zinc finger nuclease knockout specific target site of myostatin gene
A DNA molecule and encoding technology, applied in genetic engineering, plant genetic improvement, and microbial-based methods, can solve problems such as heavy experimental workload, low success rate, and long cycle time, and achieve improved research efficiency and gene knockout high efficiency effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0027] Example 1. Design and Construction of ZFN Plasmid for Specific Target Site Sequence of Porcine MSTN Gene
[0028] 1. Design of specific target site sequence for porcine MSTN gene
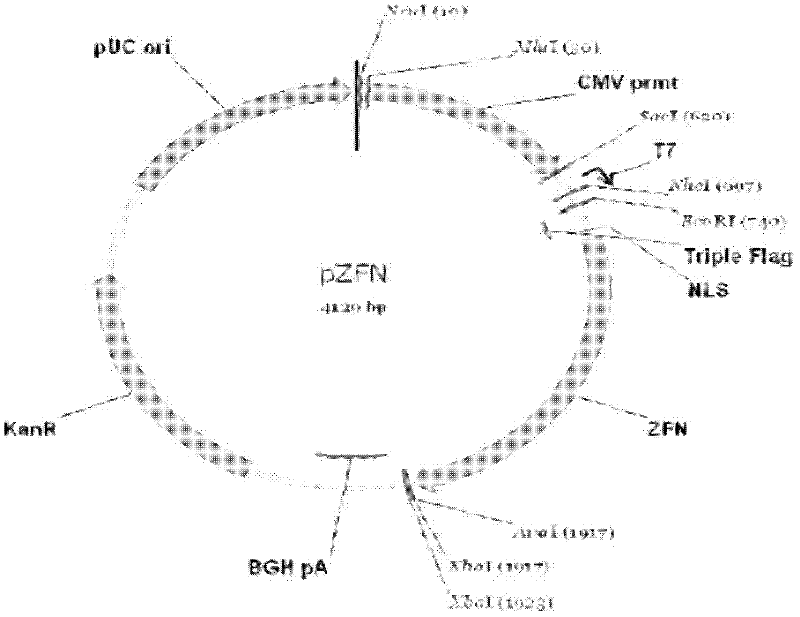
[0029] Construct a zinc liponuclease (ZFN) plasmid pZFN plasmid that knocks out specific sites in the target sequence of the Meishan pig MSTN gene. The ZFN is composed of a DNA binding domain and a DNA cutting domain. The ZFN pair specifically recognizes and binds ≥ 24 bp bases in the coding region , to ensure the high precision and high efficiency of ZFN cutting MSTN gene.
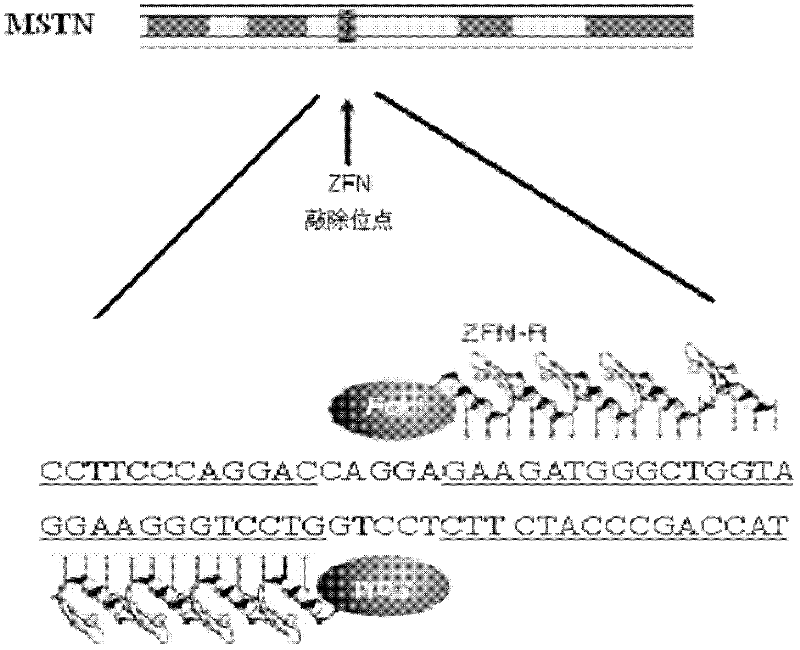
[0030] Zinc lipid nuclease (ZFN) specific knockout target site sequence is located in the second exon of MSTN gene (the nucleotide sequence of MSTN is sequence 4), and the specific target site sequence is CCTTCCCAGGAC cagga GAAGATGGGCTGGTA (artificially synthesized, sequence 1, Sequence 1 is the 3770-3801 nucleotides from the 5' end of sequence 4), see for details figure 1 As shown, the partial sequences at both ends (...
Embodiment 2
[0034] Example 2, zinc finger nuclease site-directed knockout of MSTN gene
[0035] 1. Isolation and culture of pig fetal fibroblast primary cells:
[0036] (1) Select purebred Meishan pregnant sows (Taicang Breeding Pig Farm) who have been pregnant for 35 days as experimental animals, take out the uterus containing the embryos through surgery, place them in a sterile beaker, seal with a parafilm and bring them into the laboratory;
[0037] (2) Remove the capsule around the embryo in a sterile ultra-clean bench, and place the embryo in a sterile beaker with PBS added;
[0038] (3) Transfer the embryos one by one to a small sterile petri dish, remove the head, tail and limbs of the embryo with sterile scissors and forceps, and cut the tissue into 1mm 3 sized pieces, and add a little serum to moisten the tissue;
[0039] (4) Spread the small pieces evenly on the wall of the bottle, and the distance between each small piece is about 0.2cm-0.5cm. After the small pieces are coate...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com