Method for identifying camellia oleifera cultivar and special primer and kit thereof
A kit and variety technology, applied in the direction of DNA / RNA fragments, recombinant DNA technology, etc., can solve the problems of allelic site length polymorphism among varieties, and achieve reliable detection results, good practicability, and simple operation
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0024] The microsatellite primers were developed based on a large number of Camellia oleifera genome sequences obtained by next-generation high-throughput sequencing. First, high-throughput sequencing was used to complete the "Shotgun" sequence determination of 348 million bases in the Camellia oleifera genome. The number of chromosomes in the diploid genome of Camellia oleifera is 2n=30, and the genome size is about 3800Mb. However, polyploidy of Camellia oleifera is common in cultivated varieties. In order to improve the efficiency of sequencing and assembly, diploid Camellia oleifera is preferably used for DNA sequencing. Zhejiang safflower camellia is diploid, and the sequencing individual selected in this patent is a Zhejiang safflower camellia collected from Taining, Fujian.
[0025]In order to meet the quality requirements of DNA sequencing, the sequencing DNA template was extracted with Qiagen DNeasy Plant Mini Kit, the concentration was detected by a micro-nucleic aci...
Embodiment 2
[0033] The 10 pairs of primers screened in Example 1 were used to detect the genotypes of 128 Camellia oleifera varieties collected from Guangxi, Hubei, Hunan, Jiangxi, Anhui, Fujian and other places. The specific method is as follows:
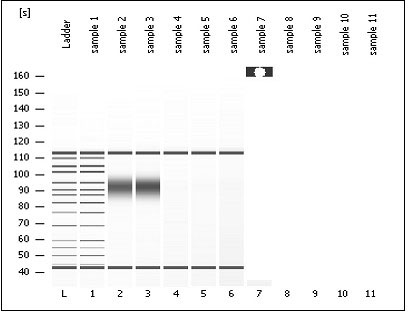
[0034] DNA was extracted from tea leaves according to the method of Murry and Thomson (1990). Then, using this as a template, 10 pairs of primers obtained by screening were used for PCR amplification on an ABI-9700 thermal cycler. The total reaction system of PCR is 15uL, and the reaction system includes: template DNA 25ng, upstream and downstream primers 20ng each (FAM label), 200uM dNTP, 0.5U Taq polymerase, 1.5uL 10×buffer (containing 100uMTris-HCl, pH 8.3, 500mM KCl, 20mM MgCl 2 and 10.0g / L BSA), add sterilized deionized water to supplement the reaction volume to 15uL. The PCR reaction conditions were: pre-denaturation at 94°C for 2 min; then denaturation at 94°C for 30 s, annealing at 50°C for 30 s, and extension at 72°C for 1 min, for...
Embodiment 3
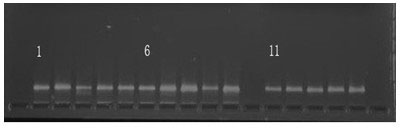
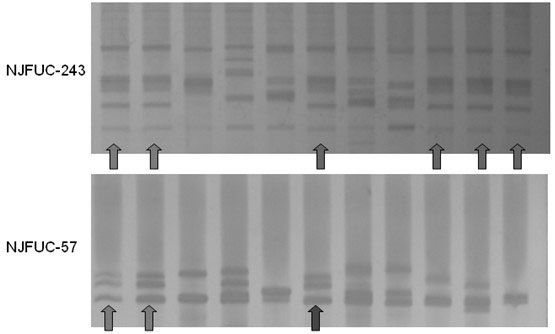
[0046] Using the primers screened in Example 1, a batch of samples sent by the Southern Seed Inspection Center of the State Forestry Administration that may be Cenruan No. 3 or may be counterfeit Cenruan No. 3 were tested, and a total of 9 samples were sent for inspection. During the experiment, the standard Cenruan No. 3 was used as the control. In order to verify the repeatability of PCR, two replicates were set up as the control. The 11 samples including the control were optionally amplified by PCR with primers NJFUC-243 and NJFUC-57. The genotypes amplified by these two primers in Cenruan No. 3 had a low frequency in the 128 varieties listed in Table 2.
[0047] In this experiment, the amplification reaction system and amplification conditions were the same as in Example 1. After the amplification, the amplified products were detected by electrophoresis using 8% polyacrylamide gel. The specific operation steps are as follows: prepare 8% polyacrylamide gel; after polymeri...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com