Specific primer pair for assisting booklice identification and application thereof
A specific primer pair and auxiliary identification technology, which is applied in the determination/inspection of microorganisms, biochemical equipment and methods, DNA/RNA fragments, etc., can solve the problem of high technical requirements for morphological identification, difficult and difficult morphological identification and other problems, to achieve the effect of simple operation, short time consumption, improved efficiency and accuracy
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0057] Example 1. Design of specific primer pairs
[0058] Based on the mitochondrial cytochrome oxidase subunit I barcode (mtDNA COI barcode) segment of ten common stored-grain booklices of the genus Booklice, we designed and synthesized specific primer pairs that can identify booklice:
[0059] Upstream primer (sequence 1 in the sequence list): 5'-GAACAATAATCGGGAGAGGA-3'; Tm=58.0°C, GC%=45.0;
[0060] Downstream primer (sequence 2 in the sequence listing): 5'-GTAGAAGAATTGCTGTAATAAG-3'; Tm=52.8°C, GC%=31.8.
Embodiment 2
[0061] Example 2. Application of specific primer pairs to identify book lice
[0062] Perform the following experiments on 22 samples:
[0063] 1. Extract the genomic DNA of single-headed book lice.
[0064] 2. Using the genomic DNA of step 1 as a template (water as a negative control), PCR amplification is performed with the specific primer pair designed in Example 1 to obtain PCR amplification products.
[0065] PCR reaction system (26ul): 2×Taq PCR Master Mix 13ul, upstream primer (10uM) 0.5ul, downstream primer (10uM) 0.5ul, genomic DNA 2ul, ddH 2 O 10ul.
[0066] PCR reaction parameters: 95°C pre-denaturation 3min; 95°C denaturation 30sec, 50°C annealing 40sec, 72°C extension 1min, 32 cycles; 72°C 8min; 4°C storage.
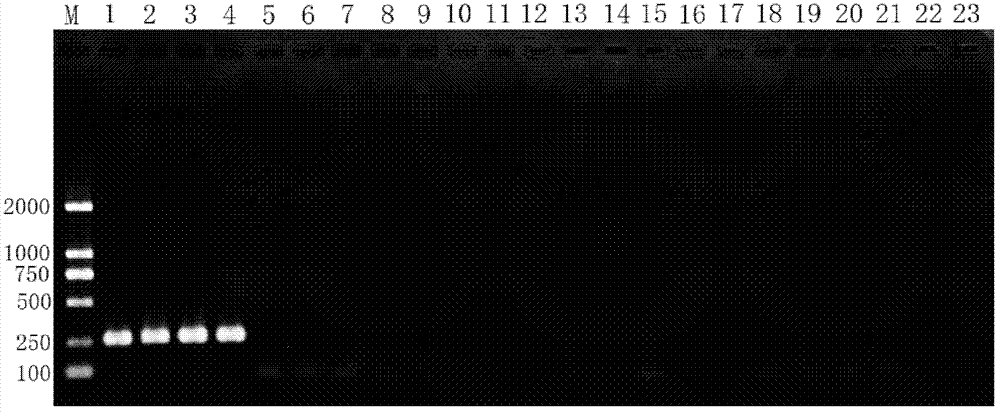
[0067] 3. Carry out 4ul of the PCR amplification product of step 2 on 1.5% agarose gel electrophoresis, stain with ethidium bromide (EB), observe and image analysis in the gel system.
[0068] See the electrophoresis results of each sample figure 1 . figure 1 In, M: DNA...
Embodiment 3
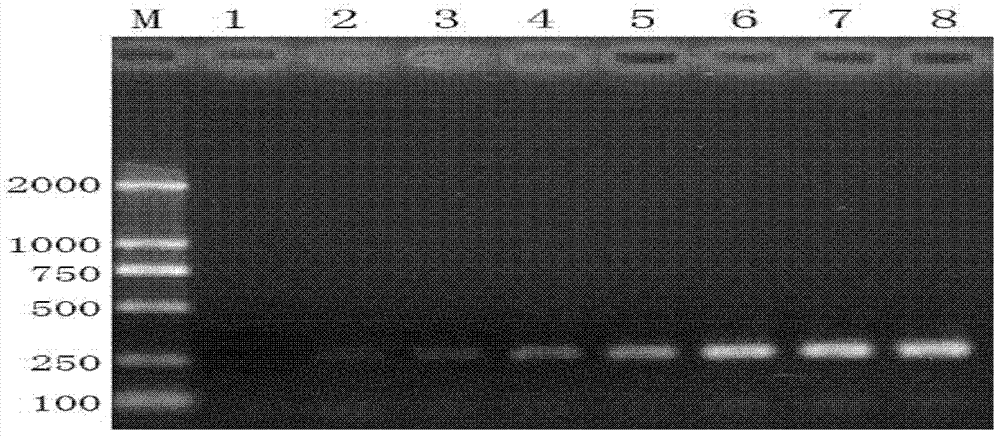
[0070] Example 3. Sensitivity detection of specific primer pairs
[0071] Experiment with sample 1 as follows:
[0072] 1. Extract the genomic DNA of single-headed book lice.
[0073] 2. Detect the concentration of genomic DNA and serially dilute it with TE buffer to obtain 8 dilutions; the concentrations of genomic DNA in the 8 dilutions are: 0.1ng / ul, 1ng / ul, 5ng / ul, 10ng / ul, 25ng / ul, 50ng / ul, 75ng / ul and 100ng / ul, which are diluent 1 to diluent 8.
[0074] 3. Using each dilution as a template, PCR amplification was performed with the specific primer pair designed in Example 1 to obtain PCR amplification products.
[0075] PCR reaction system (26ul): 2×Taq PCR Master Mix 13ul, upstream primer (10uM) 0.5ul, downstream primer (10uM) 0.5ul, diluent (containing genomic DNA) 1ul, ddH 2 O 11ul.
[0076] PCR reaction parameters: 95°C pre-denaturation 3min; 95°C denaturation 30sec, 50°C annealing 40sec, 72°C extension 1min, 32 cycles; 72°C 8min; 4°C storage.
[0077] 4. Perform 4ul of the PCR...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com