Kit for detecting drug-resistant site of influenza viruses A (IFVA), preparation thereof and application thereof
A technology of influenza A virus and detection kits, applied in the determination/testing of microorganisms, biochemical equipment and methods, fluorescence/phosphorescence, etc., can solve the problems of long operation cycle, inconvenient hospital development and promotion, expensive detection equipment, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0049] The preparation of embodiment 1 kit
[0050] 1) Preparation of IFVA-H274Y nucleic acid fluorescent PCR detection mixture:
[0051] Mix IFVA upstream primer 0.75 μl / test; downstream primer 0.75 μl / test; probe 0.05 μl / test; RT-PCR MIX 12.5 μl / test; liquid.
[0052] Upstream primer: 5'-tggacaggcctcatacaaga-3' (SEQ ID NO: 2)
[0053] Downstream primer: 5'-attcgagccatgccagttat-3' (SEQ ID NO: 3)
[0054] Probe: 5'-FAM-CCtCAtaGtAAtaa-BHQ1-3' (SEQ ID NO: 4)
[0055] The above primers and probes were synthesized by conventional methods.
[0056] 2) The IFVA-H274Y fluorescent PCR detection mixture, RT-PCR enzyme, positive control substance and negative control substance are individually packaged and assembled into a kit.
[0057] Positive control substance: H274Y mutant strain constructed by genetic engineering
[0058] The preparation method of positive control substance is:
[0059] Use the above upstream and downstream primers (SEQ ID NO: 2 and SEQ ID NO: 3) to amplify ...
Embodiment 2
[0065] Application of embodiment 2 detection kit
[0066] Adopt the kit prepared by embodiment 1
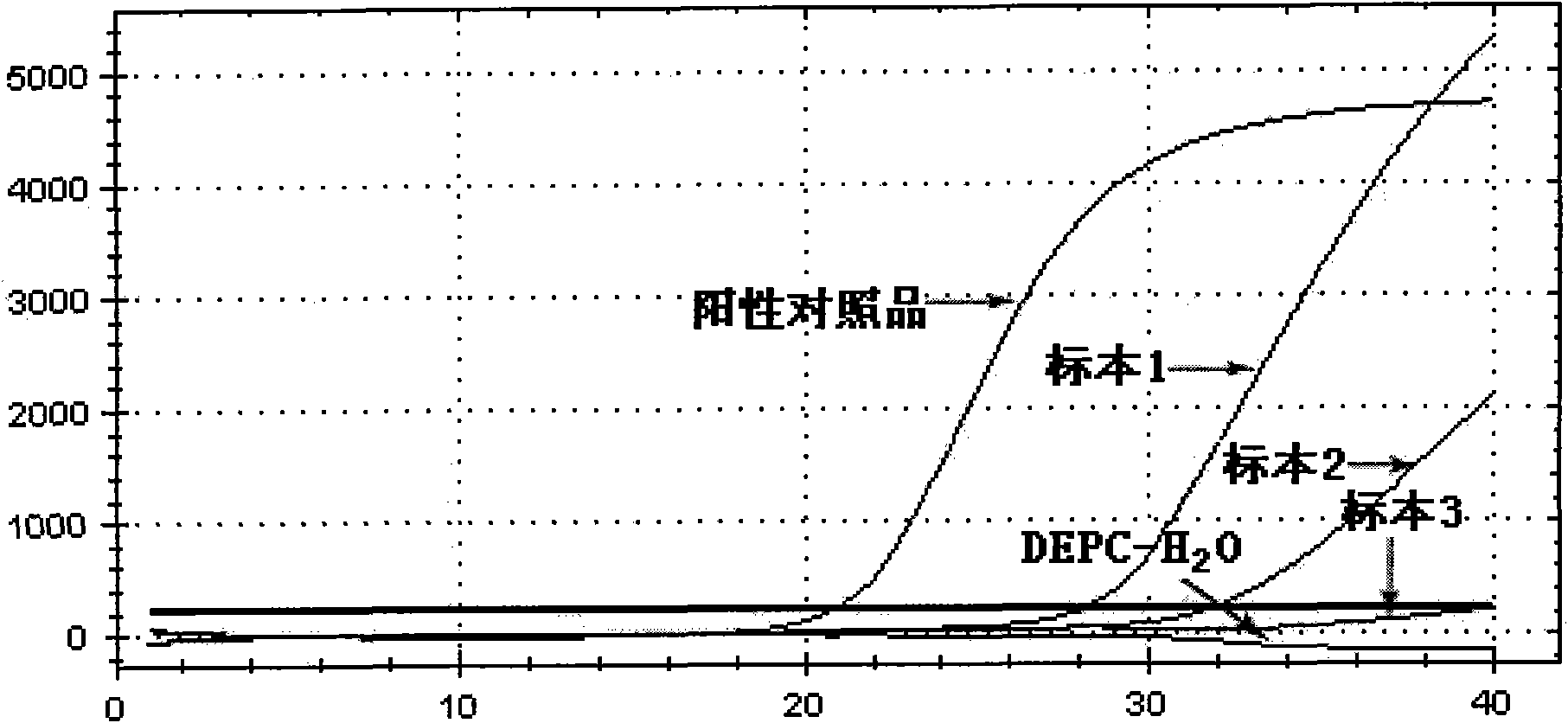
[0067] Specimen 1, 2, and 3 were obtained from clinical specimens.
[0068] 2.1 Processing of test specimens:
[0069] Take 100 μl of the sample and place it in a 1.5ml centrifuge tube, add 300 μl of lysate (Trizol), and then add 100 μl of chloroform, shake and mix on a mixer for 5 sec or invert for 15 times. Centrifuge at 13,000 rpm for 15 min. Aspirate the upper layer liquid, add an equal volume of isopropanol, invert and mix well, and let stand at room temperature for 10 minutes. Centrifuge at 13,000 rpm for 10 min, pour off the supernatant; add 700 μl of 75% ethanol, and wash upside down. Centrifuge at 13,000rpm for 5min, place it upside down on absorbent paper, and discard the liquid. Centrifuge at 4,000rpm for 5sec, shake the residual liquid on the tube wall to the bottom of the tube, use a micro-sampler to absorb the liquid as much as possible, add 30μl DEPC-H 2 O di...
Embodiment 3
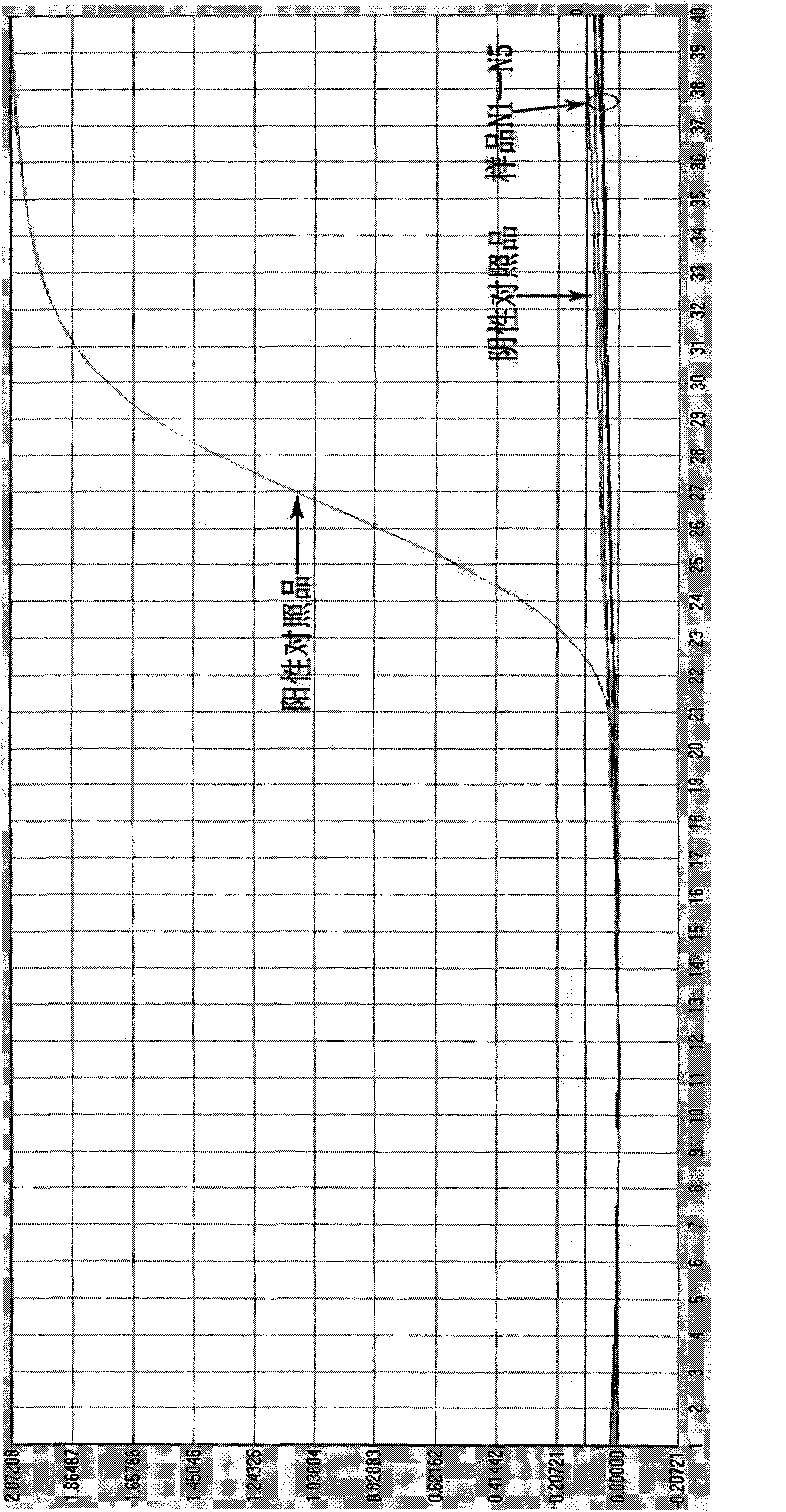
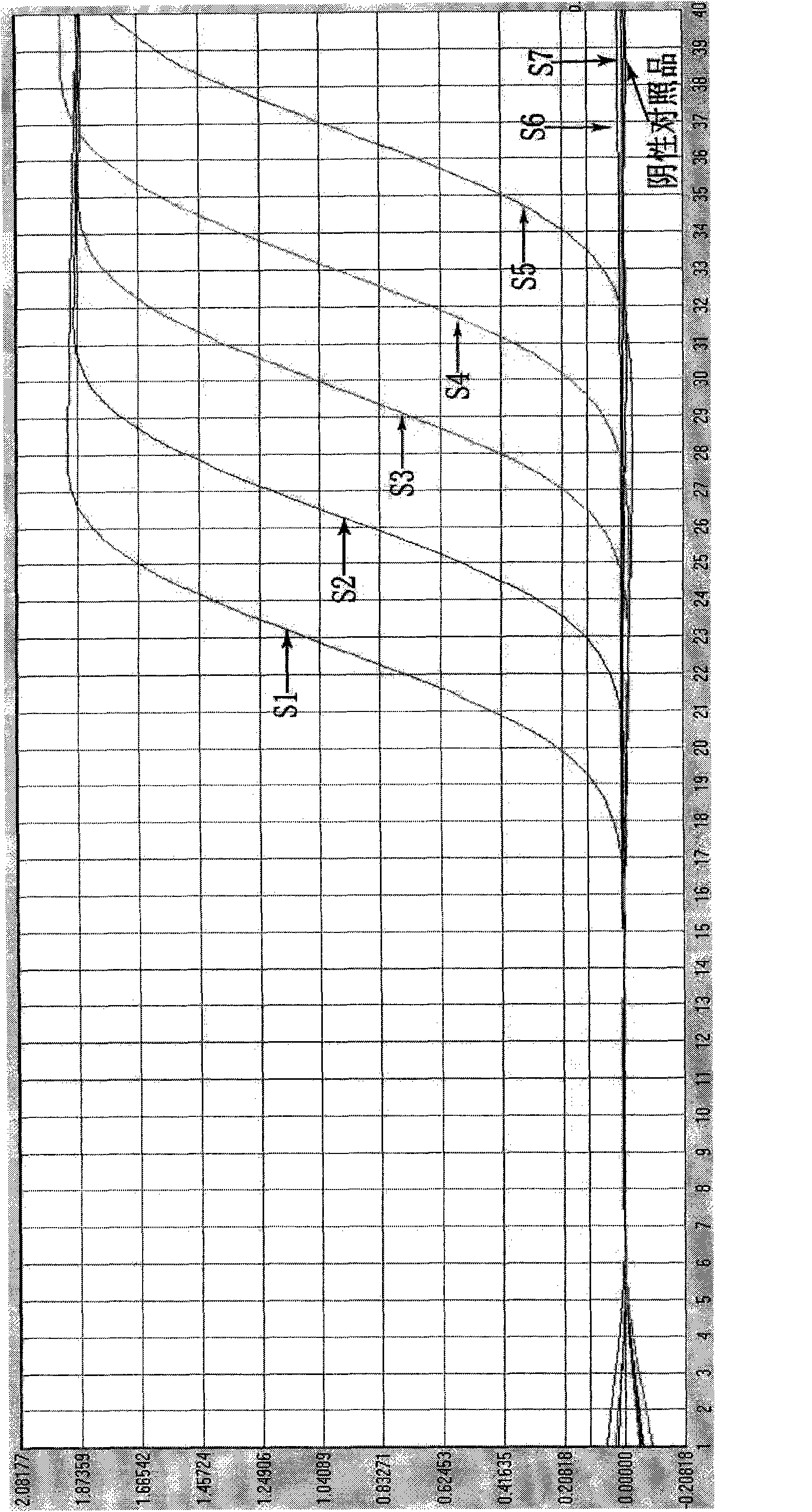
[0084] The specificity research of embodiment 3 detection kits
[0085] Adopt the kit prepared by embodiment 1
[0086] Influenza A virus H1 subtype specimens, influenza A virus H3 subtype specimens, influenza B virus specimens, enterovirus 71 specimens obtained clinically; negative cell culture supernatants of influenza A H1N1 influenza virus culture cell line MDCK 1 serving. The sample numbers are N1, N2, N3, N4 and N5, respectively.
[0087] 3.1 Processing of test specimens:
[0088] Take 100 μl of the sample and place it in a 1.5ml centrifuge tube, add 300 μl of lysate (Trizol), and then add 100 μl of chloroform, shake and mix on a mixer for 5 sec or invert for 15 times. Centrifuge at 13,000 rpm for 15 min. Aspirate the upper layer liquid, add an equal volume of isopropanol, invert and mix well, and let stand at room temperature for 10 minutes. Centrifuge at 13,000 rpm for 10 min, pour off the supernatant; add 700 μl of 75% ethanol, and wash upside down. Centrifuge a...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com