Preparation method and application of gene chip for determination and drug resistance detection of influenza A virus
A type of influenza A virus, gene chip technology, applied in the field of gene chip detection, can solve the problems of long time, no specificity and sensitivity, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0047] Example 1: Development of Influenza A Virus Nucleic Acid Screening and Drug Resistance Detection Gene Chip
[0048] 1. General primer design and screening
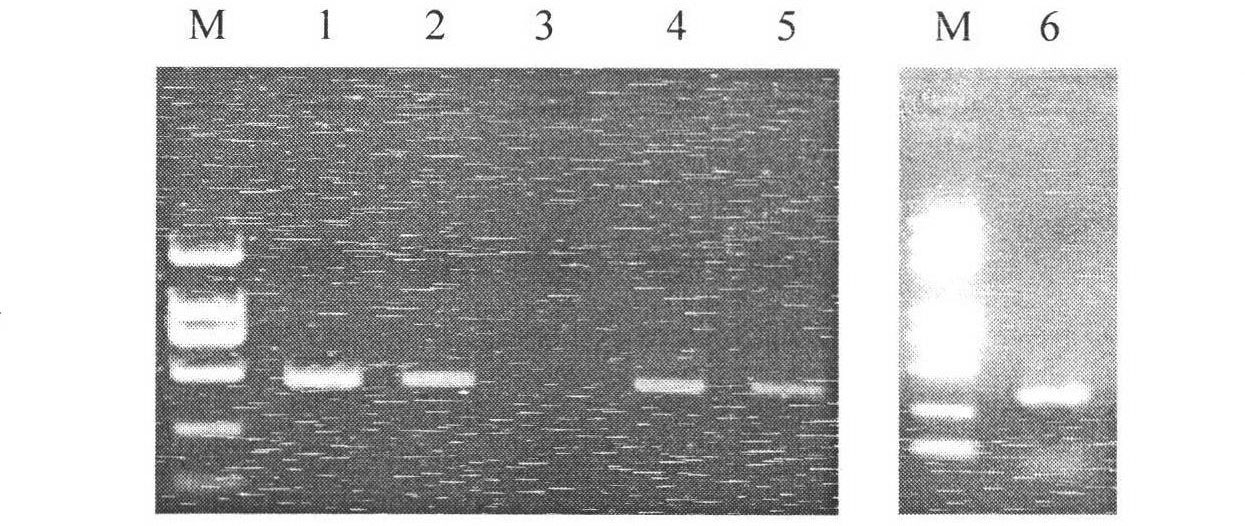
[0049] In order to achieve simultaneous universal amplification of 4 subtypes of N1 influenza virus and 1 H3N2 influenza virus, 2 pairs of primers were used to amplify 4 N1 subtype viruses, and 1 pair of primers was used to amplify H3N2. The primer sequences and corresponding target See Table 1 for virus information. The combination of NF11 / NF12 and NR21 / NF22 amplified four N1 subtypes, the amplified fragment was 452bp, including the drug resistance mutation site H274Y; NF5 and NR5 amplified H3N2, and the amplified fragment was 281bp, including the drug resistance mutation site Point E119V. The agarose electrophoresis results of the amplified products are shown in the appendix figure 1 .
[0050] 2. Screening of specific probes
[0051] The screening of typing probes first excludes the non-specific crossover be...
Embodiment 2
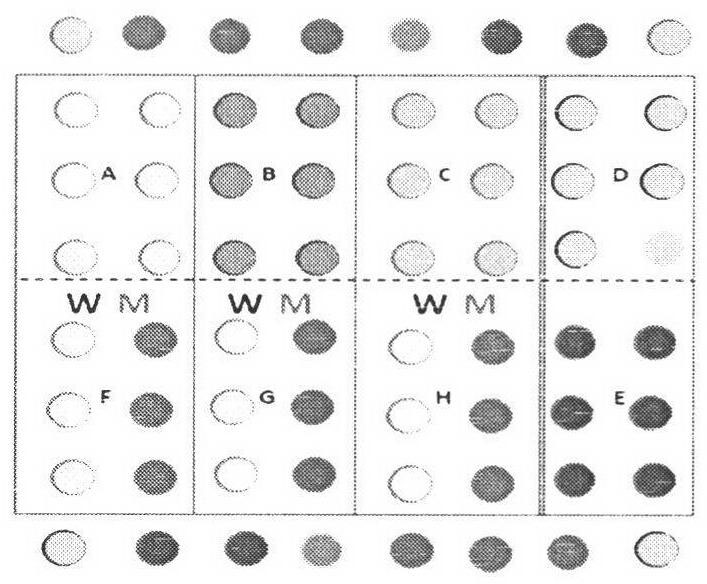
[0059] Example 2: Determination of Gene Chip Positive Judgment Criteria
[0060] The cutoff value is a criterion for judging whether the signal value of the gene chip is positive. Each typing probe selects non-influenza virus (i.e., negative strain) and blank control for gene chip hybridization, and through repeated experiments and data statistics, the background statistical average value +2SD of negative strain and blank control is used as each The cutoff value of the probe is shown in Table 6. The discrimination ability of each mutation detection probe is more than 2.5 times as the criterion for judging whether there is a mutation at the drug resistance site.
[0061] Table 6 Determination of the Cutoff value of each probe
[0062]
Embodiment 3
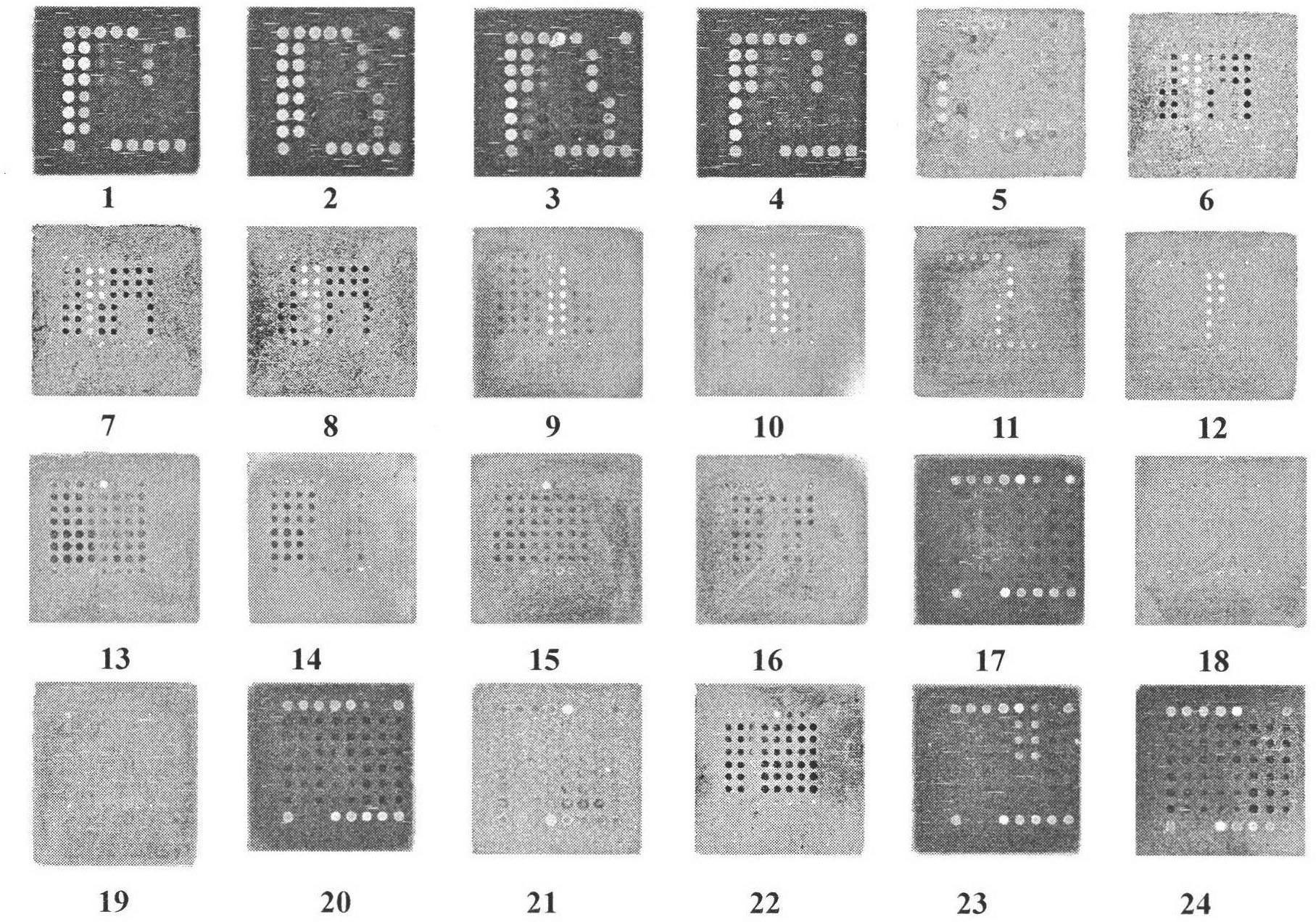
[0063] Example 3: Influenza A virus nucleic acid screening and gene chip specificity evaluation for drug resistance detection
[0064] Specificity is the most important assessment index of a diagnostic method. Using the optimized system and conditions, the gene chip of the present invention has detected 24 strains of various types of influenza viruses, parainfluenza viruses, and common respiratory viruses. The test results are shown in the attached image 3 . As can be seen from the figure, the present invention can not only correctly distinguish influenza A virus from other common respiratory tract viruses and influenza B virus, but also distinguish influenza A H1N1, seasonal H1N1, and seasonal H3N2 among influenza A viruses. It can be correctly distinguished from avian influenza H5N1 with good specificity. In addition, a seasonal H1N1 influenza virus Tamiflu resistance mutation site was found to be mutated in the experiment. After sequencing verification, the result was con...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Titer | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com