Cloning and application of arabidopsis oidium disease resistance suppressor gene EDTS3
A technology for suppressing genes and Arabidopsis thaliana, applied in the fields of application, genetic engineering, plant genetic improvement, etc., can solve the problem of low resistance and achieve the effect of improving disease resistance
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0034] Example 1 Analysis of the edts3 mutant suppressing disease resistance phenotype
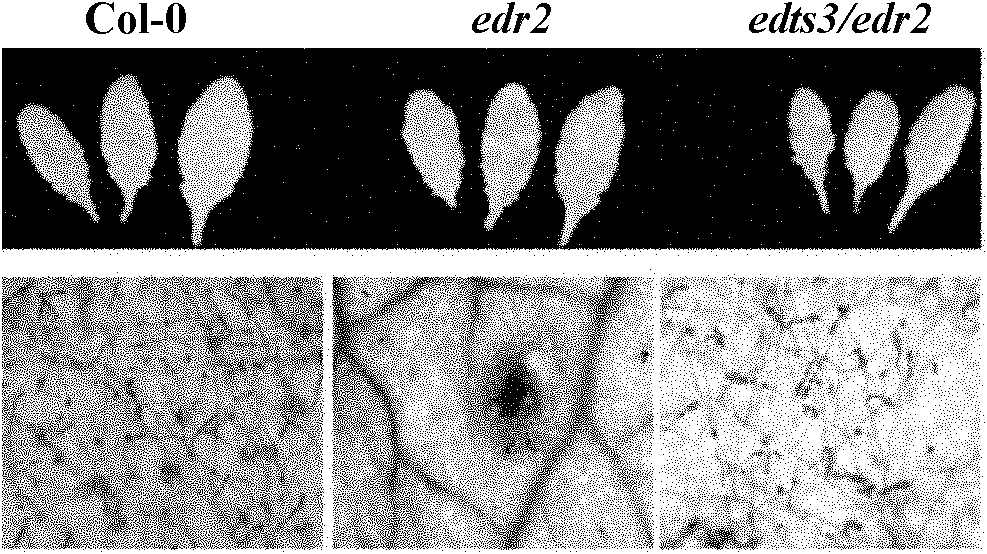
[0035] 1. The edts3 / edr2 mutant inhibits the powdery mildew resistance and powdery mildew-induced cell death of the edr2 mutant
[0036] (1) Materials and methods
[0037] edts3 / edr2 (wherein edts3 is enhanced disease resistance two suppressor 3) is a mutant edr2 (4) that can suppress edr2 powdery mildew bacteria obtained by screening from the mutagenized population by using 0.03% EMS (purchased from sigma company) Mutants with enhanced resistance. Then a mutant was named edts3 / edr2.
[0038] First, the seeds of mutant edr2, edts3 / edr2 and wild-type col-0 (purchased from Arabidopsis Biological Resource Center (ABRC)) were sown on MS medium (commercially purchased from PhytoTechnology Laboratories), and vernalized at 4°C for 2-3 days , transferred to a plant growth chamber under 9h light / 15hdark light conditions. About 7-10 days, move the seedlings to the soil. After 4-6 weeks of growt...
Embodiment 2
[0041] Embodiment 2 Obtaining of EDTS3 gene
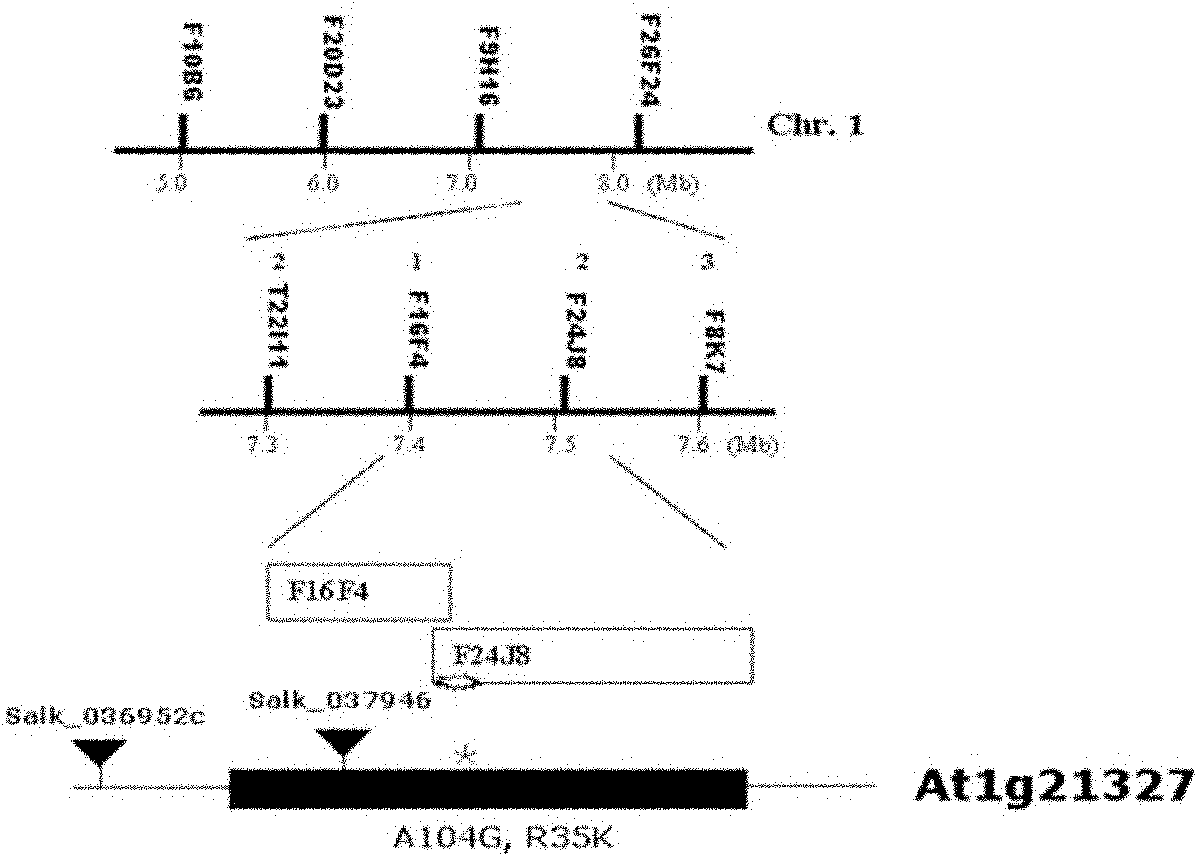
[0042] 1. Isolation of EDTS3 gene by map-based cloning
[0043] We isolated the EDTS3 gene using map-based cloning. The specific method is: the mutant edts3 / edr2 is crossed with the ecotype Lansberg (commercially purchased from ABRC), and the obtained F1 generation is self-crossed to generate the F2 generation. In the F2 generation, a single plant with a wild-type phenotype of the edr2 mutation was selected and then selfed to generate the F3 generation. Wild-type individuals were selected from each line of the F3 generation, and by PCR method and using chromosome positioning markers evenly distributed on the five chromosomes (http: / / signal.salk.edu / genome / SSLP_info / SSLPordered.html), the The gene was initially located at the front end of the broken arm of the first chromosome. Subsequently, about 3000 F3 generation individual plants were used to fine-map the gene in the 40kb region between F9H16 and F26F24 (8)( figure 2 ). Th...
Embodiment 3
[0046] Example 3 Functional Verification of EDTS3 Gene
[0047] In order to verify whether the resistance inhibition of edts3 mutation to edr2 mutant powdery mildew is caused by the mutation of At1g21327 base, we constructed a genetic transformation vector of At1g21327 with its own promoter, and transformed double mutant edts3 / edr2 plants by flower infection , in the T1 generation, the transgenic line recovered the phenotype of the edr2 mutant.
[0048] We used the wild-type col-0 DNA as a template, and used a pair of PCR primers F: 5′-AACTGCAGAAGTGGTGCTCACGTTGTTGTATTT-3′ and R: 5′-AACTGCAGAAGTGGTGCTCACGTTGTTGTATTT-3′ to amplify the 5.7kb At1g21327 gene with KpnI and PstI fragment.
[0049] PCR reaction system
[0050] DNA: 2.0ul
[0051] 10xbuffer: 5.0ul
[0052] dNTPs (2.0mM): 5.0ul
[0053] MgSO4 (25mM): 2.0ul
[0054] Primer F (10uM): 1.6ul
[0055] Primer R (10uM): 1.6ul
[0056] KOD Taq (1U): 1.0ul
[0057] H2O: 31.8ul
[0058] PCR reaction program: pre-denatur...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com