A group of oligonucleotide sequences capable of identifying Vibrio harveyi and Vibrio alginolyticus synchronously and preparation method of the oligonucleotide sequences
A technology of Vibrio harveii and Vibrio alginolyticus, applied in the field of oligonucleotide sequences, can solve the problems of easy generation of drug-resistant strains, false positives, long cycle and the like
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0083] The preparation steps of a set of oligonucleotide sequences capable of simultaneously identifying Vibrio harveyi and Vibrio alginolyticus in this embodiment mainly include:
[0084] 1. Preparation of Vibrio liquid
[0085] Wash the colony of Vibrio harveyi and Vibrio alginolyticus from the slope with normal saline, centrifuge at 6000rpm, discard the supernatant, resuspend and dilute the bacterial solution with normal saline to the following concentration range: 1×10 8 ~9×10 8 pc / ml, stored at -20°C for later use;
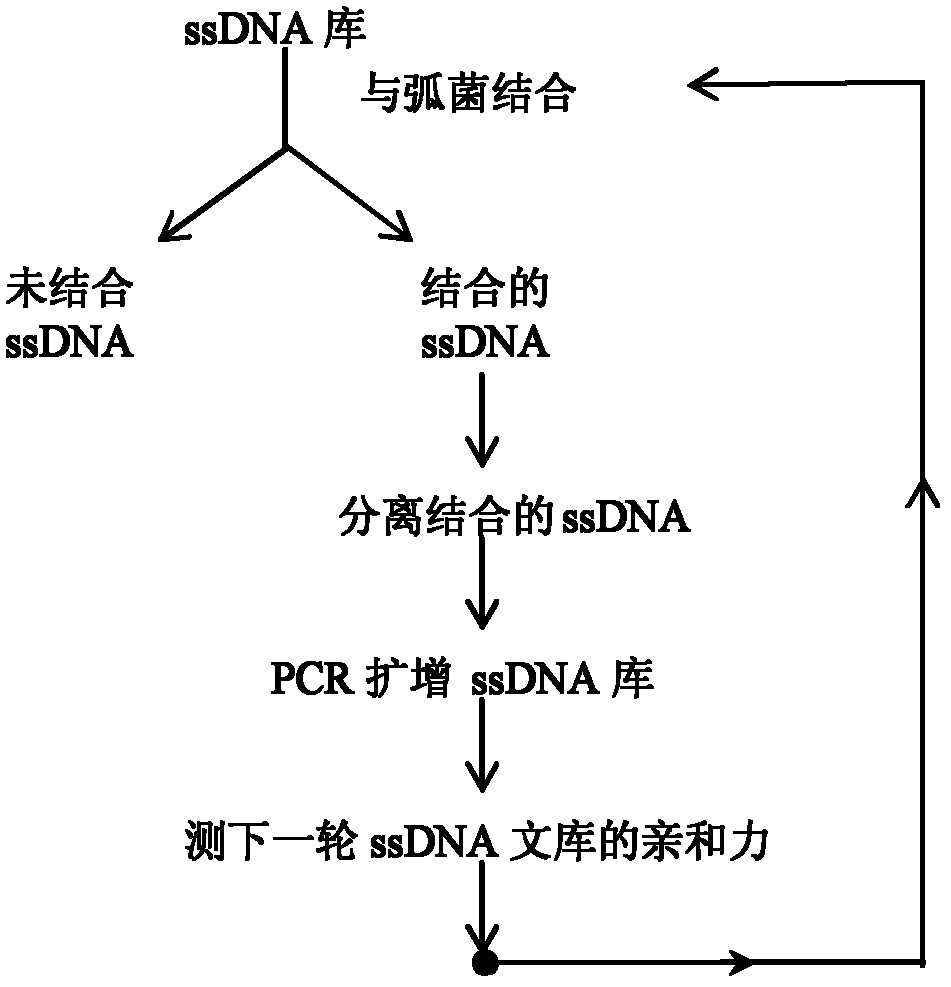
[0086] 2. SELEX screening of aptamers
[0087] 2.1 Combination and isolation of ssDNA with Vibrio harveyi
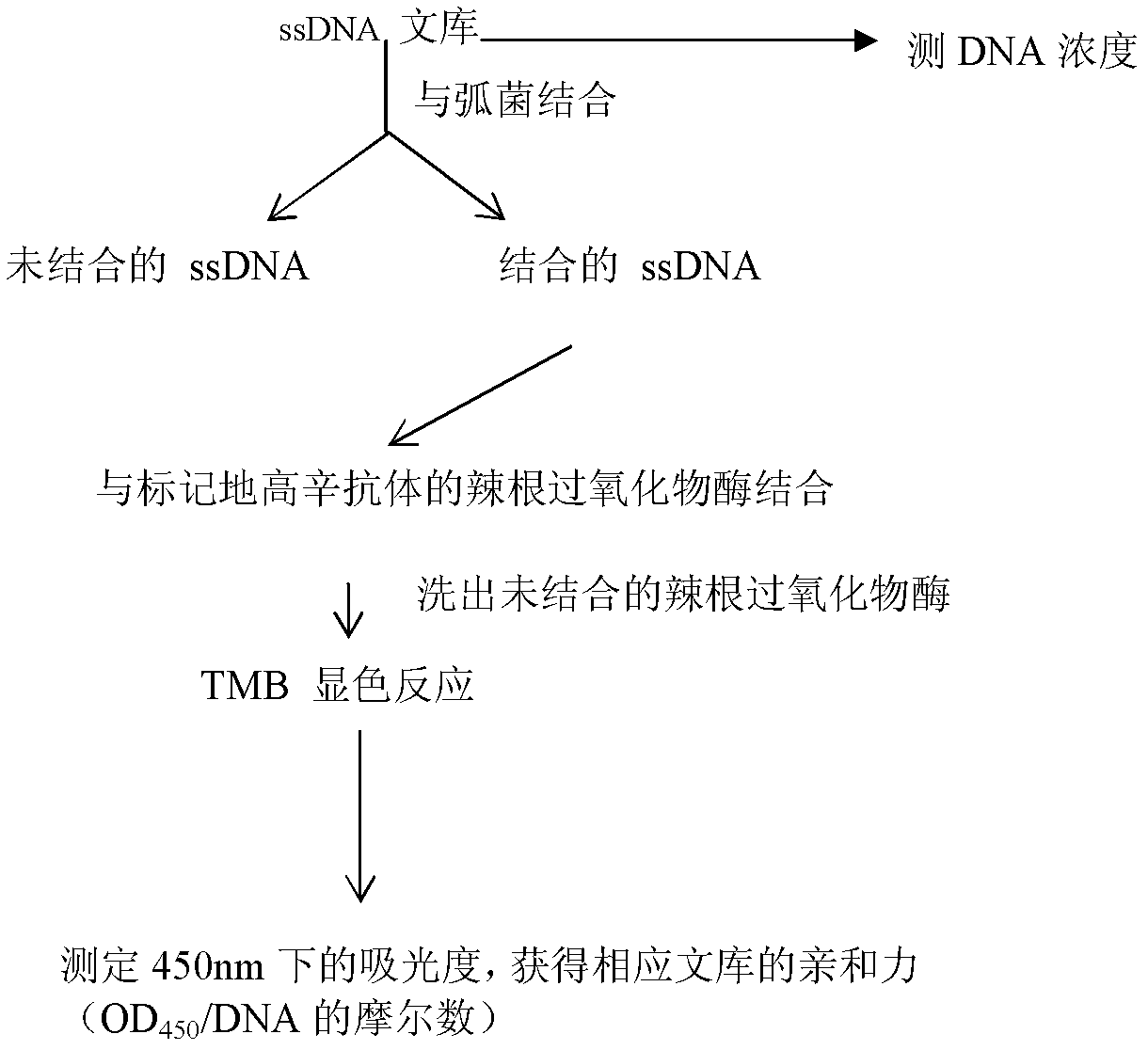
[0088] Take 4 μL of 100 μM ssDNA oligonucleotide library, dilute to 100 μl with 2× binding buffer, denature at 95°C for 5 minutes, add 100 μl of Vibrio harveii bacteria solution restored to natural room temperature after 10 minutes in ice bath, combine on a shaking table for 30 minutes, and then Centrifuge at 6000rpm for 5min, discard the supernatan...
Embodiment 2
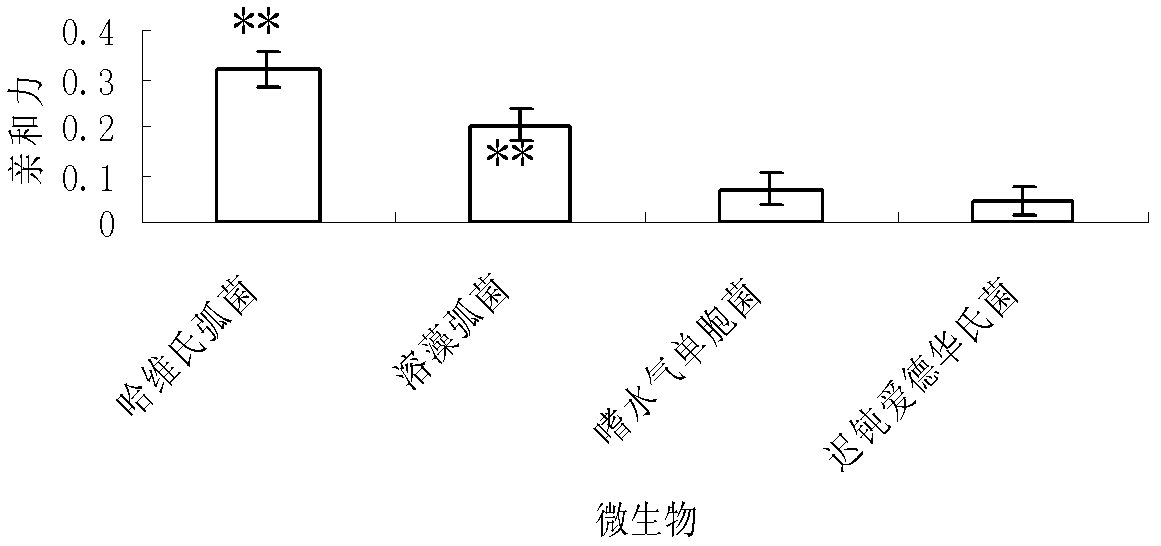
[0131] The oligonucleotide sequence has the purpose of identifying and detecting Vibrio harveyi: the specific steps are as follows: 1) get the wound tissue mucus of the diseased fish, dissolve it in distilled water and inoculate it in the tryptone soybean broth medium (TSB) medium, Incubate on a shaker at 100 rpm at 30°C for 8-10 hours. Then take the bacterial solution and centrifuge, discard the supernatant culture solution, then dilute it with normal saline to 1×108~9×108 cells / mL, and store it at -20°C for later use. Then carry out affinity detection according to the method of the affinity-specific verification of the step 4.3 in the embodiment 1 with the aptamer C7 (SEQ ID No.1) that 5 ' end is marked with digoxin; Alginolyticus and Vibrio alginolyticus were used instead of the test bacteria solution, and the affinity test was carried out in the same way to obtain two positive controls; sterile distilled water was used instead of the test bacteria solution, and the affinit...
PUM
| Property | Measurement | Unit |
|---|---|---|
| absorbance | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com