Method for qualitatively and quantitatively detecting nucleic acid based on high-flux sequencing technology
A quantitative detection, high-throughput technology, applied in the direction of microbial determination/inspection, biochemical equipment and methods, etc., to achieve the effect of simple operation and less sample volume
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
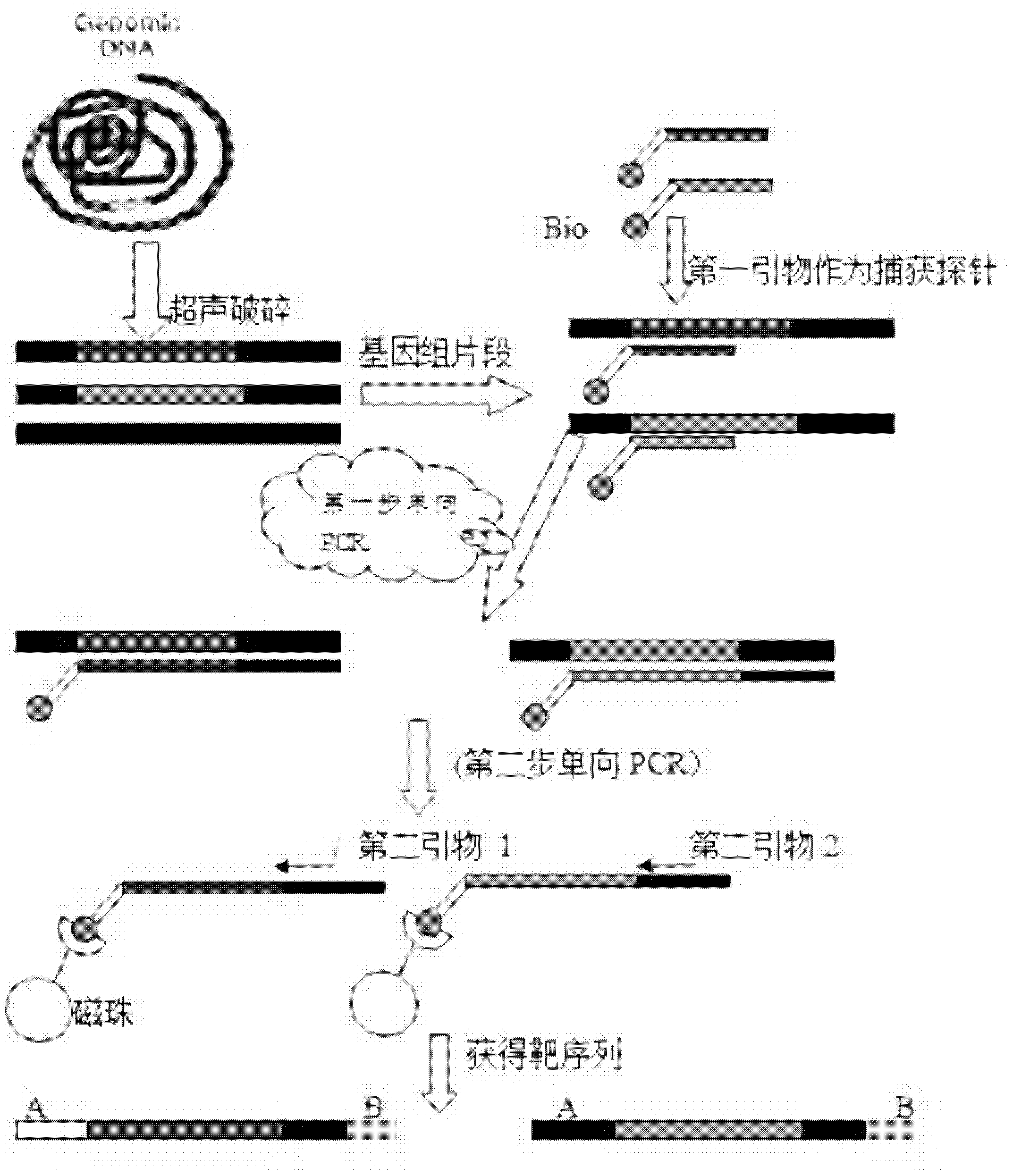
[0085] Example 1 Example of a method for capturing specific nucleic acid targets by two-step unidirectional amplification of multiple genes and multiple regions
[0086] 1. Genomic DNA extraction and purification
[0087] 1. Add 400 μl Lysis Solution and 20 μl Proteinase K solution to 200 μl whole blood, mix well to obtain a homogeneous suspension.
[0088] 2. Incubate in a 56°C water bath for 10 minutes.
[0089] 3. Add 200 μl absolute ethanol and mix well.
[0090] 4. Transfer the prepared lysis mixture to the Gene JET Genomic DNA Purification Column. Centrifuge at 6000g for 1 min and discard the waste liquid. Transfer the Gene JET Genomic DNA Purification Column to a new 2ml collection tube.
[0091] 5. Add 500μl Wash buffer I, centrifuge at 8000g for 1min, discard the waste liquid, and put the purification column back into the collection tube.
[0092] 6. Add 500μl Wash buffer II to the Gene JET Genomic DNA Purification Column, and centrifuge at 12000g for 3min.
[0...
Embodiment 2
[0140] Example 2 Non-invasive (non-invasive) prenatal screening diagnosis of Down's syndrome and various congenital genetic diseases
[0141] Chromosomal abnormalities in the baby's cells can cause disease. There are 23 pairs or 46 chromosomes in normal human cells, but fetuses with chromosomal abnormalities will have increased chromosomes or decreased chromosomes. It leads to various diseases after birth. For example, Down syndrome with a high incidence is caused by three chromosomes 21. At present, if it is necessary to clearly diagnose whether it is a fetus with chromosomal abnormalities, it is necessary to draw fetal blood, amniotic fluid or placental villi for examination. These methods are all invasive examinations, which have certain risks to the fetus and pregnant women. The risks of fetal miscarriage, premature delivery, and intrauterine infection are about 1%-2%. If non-invasive means can be used to identify fetuses with chromosomal abnormalities from the DNA test ...
Embodiment 3
[0166] Example 3 Sequencing Qualitative and Quantitative Sequencing of Human Microbial Species and Their Relationship with Diseases and Health
[0167] With the continuous development of life science research, human beings have entered the post-genome era. The health of the human body depends on the interaction between genes and the environment. At present, a large number of studies focus on the analysis of the relationship between human genetic makeup and disease susceptibility and drug sensitivity. However, it is not only human genes that play a role in the body and affect people's life, aging, illness and death, but also the genes of a large number of symbiotic microorganisms. The sum of their genetic information is called the microbiome. The human microbiome refers to the general term for symbiotic, commensal and pathogenic microorganisms in the ecological community existing in the human body (mainly in the intestine) or on the surface. If a human being is defined as a co...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com