New method for capturing chromatin nucleosome vacancy district at high-throughput complete genome level and use therefor
A genome-wide and chromatin technology, applied in the field of medical molecular biology, can solve problems such as low accuracy
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0026] Step 1. Lyse cells and extract nuclei
[0027] HeLa S3 cells were cultured to reach 80% confluency and replaced with serum-free medium. After 24 hours of serum starvation [15, 16], the culture medium was discarded. Wash the cells twice with ice-cold PBS, then add 1ml of PBS, scrape the cells on ice with a cell scraper, centrifuge at 3000rpm for 10 minutes at 4°C, and collect the cells. The cells were suspended with PBS buffer, and the cells were counted. 10 7 Aliquot / tube and centrifuge again to collect cells. Add nuclei isolation buffer (10mM Tris buffer, pH 7.4, 10mM NaCl, 5mM MgCl 2 , 1mM PMSF, 0.2% NP40), mix gently to make the final cell concentration 5×10 6 / ml, ice bath for 10 minutes. Centrifuge at 3000 rpm for 10 minutes at 4°C to collect cell nuclei. Nuclei were washed with NP40-free buffer (10 mM Tris buffer, pH 7.4, 10 mM NaCl, 5 mM MgCl 2 , 1 mM PMSF), washed away excess NP40, and centrifuged at 3000 rpm for 10 minutes at 4°C.
[0028] Step 2. Neutr...
Embodiment 2
[0074] Step 1. High-throughput sequencing of NFR library
[0075] 1. Preparation of sequencing samples
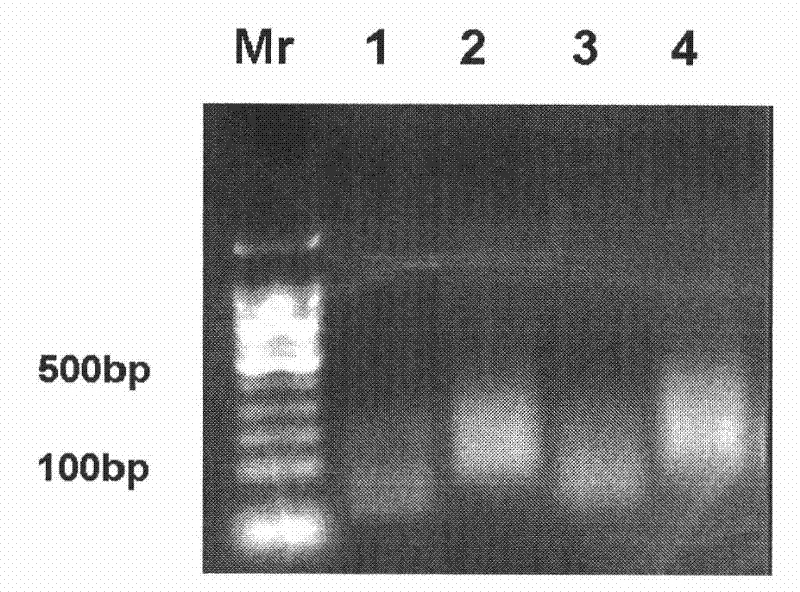
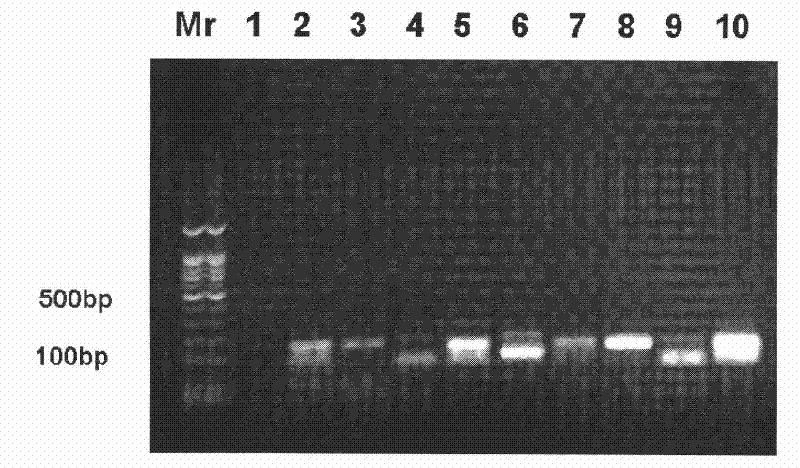
[0076] The NFR library preparation method is the same as the above step 1 to step 8, extract HeLaS3 nucleus; neutral formaldehyde fixes the nucleus; DNase I makes nick; nick DNA translation biotin incorporation; S1 nuclease cut nick DNA; phenol / chloroform extraction, ethanol precipitation; DNA A was added to the end of the fragment, and a linker was added; biotin-DNA was picked up by magnetic beads; NFR library was amplified; DNA fragments were recovered from the gel; The samples were sent to BGI and Illumina's Solexa Genome Analyzer platform for high-throughput sequencing.
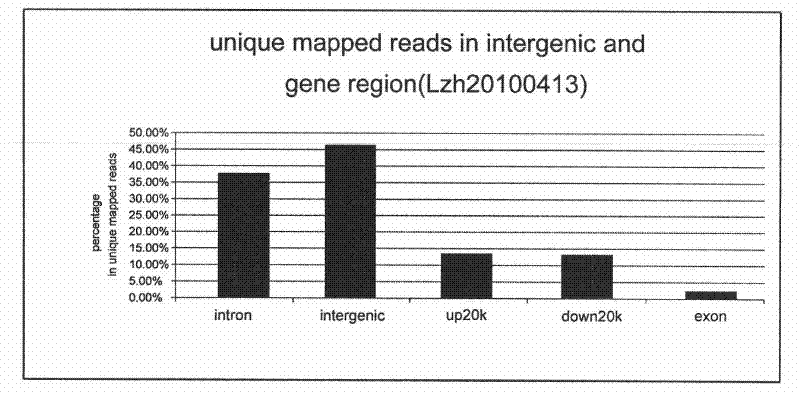
[0077] 2. The distribution of Unique mapped reads in coding regions and intron regions. Use the known information of UCSC to annotate the Sequencing data, calculate the proportion of exons, introns and conserved non-coding regions in the entire Sequencing data, and compare it with the whole genome da...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com