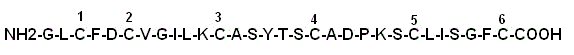Positioning method of disulfide bonds in polypeptide
A positioning method and disulfide bond technology, applied in measuring devices, material analysis through electromagnetic means, instruments, etc., can solve problems such as laborious, unstable analysis results, time-consuming, etc., and achieve simple operation, accurate results, and integrity maintenance Effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0052] In order to fully illustrate the technical solution of the present invention, an example is used to locate the connection mode of disulfide bonds in a polypeptide (molecular weight 3140) composed of 30 amino acids (6 cysteines among which form disulfide bonds with each other), and its primary structure Such as figure 1 shown.
[0053] The first step, partial reduction of the disulfide bond of the polypeptide sample to be tested
[0054]Add 100 μL of 0.5M DTT solution (pH 6.5) to 2 mg of sample and incubate for 30 min (60°C), add 10 μL of TCEP (0.2M) solution, react at 60°C for 20 min, add 50 μL of 0.2% TFA aqueous solution to terminate the reaction. Separation was carried out on 515HPLC of Waters Company in the United States, the separation column was Kromasil 250*4.6mm C18 column, and the separation gradient was as follows:
[0055] time (min) Flow rate (mL / min) A% (0.1% TFA / H 2 O) B% (0.1% TFA / H 2 O) 0 1 80 20 40 1 60 40 ...
Embodiment 2
[0063] The second step is to alkylate the uncycled cysteine sulfhydryl groups of peak A and peak B
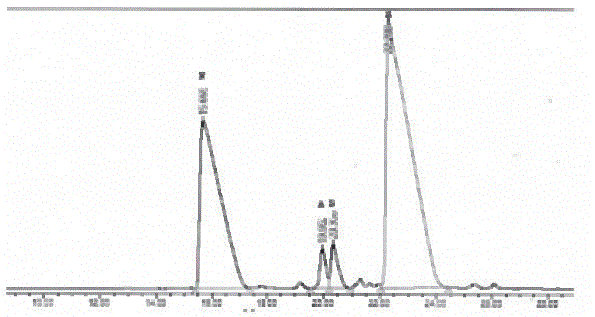
[0064] React the freeze-dried sample of peak A with 50 μl of 1.0 M iodoacetamide solution at 60°C for 10 minutes, add 100 μl of 0.2% TFA aqueous solution to terminate the reaction, and separate on 515HPLC of Waters Company in the United States. The gradient is as before, and the chromatogram is as follows Figure 7 shown.
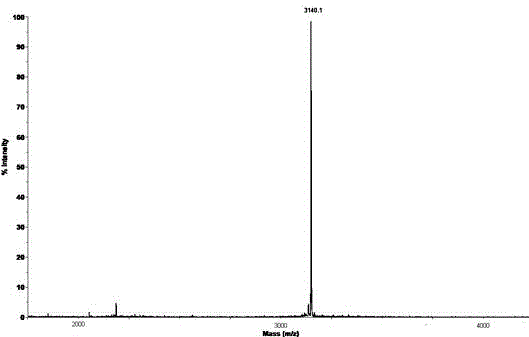
[0065] Collect 10.14min elution peak, adopt MALDI-TOF mass spectrum identification, its molecular weight is 3376 (see Figure 8 ).
[0066] The results of mass spectrometry showed that the molecular weight of the chromatographic peak with a retention time of 10.14min was 232Da more than that of peak A before being alkylated, indicating that four semi-light amino acid residues were modified by alkylation.
[0067] React the lyophilized sample of peak B with 50 μl of 1.0 M iodoacetamide solution at 60°C for 10 minutes, add 100 μl of 0.2% TFA aqueous s...
Embodiment 3
[0071] Step 3: Break all disulfide bonds in the polypeptide chain (all reduction)
[0072] The partially reduced product of the alkylation was reacted with 1M dithiothreitol (DTT) at 50°C for 20 minutes to break all the disulfide bonds in the polypeptide chain, and samples were taken for mass spectrometry identification after the reaction.
[0073] Mass spectrometry results show that the molecular weight of the alkylated A peak eluate is 3378 after the reaction, which is 2 Da more than before the reaction, indicating that a pair of disulfide bonds are broken (see Figure 11 ).
[0074] Mass spectrometry results show that the molecular weight of the alkylated peak B eluate after reaction is 3262, which is 4Da more than before the reaction, indicating that there are two pairs of disulfide bonds broken (see Figure 12 ).
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com