Species-specific primer pair for detecting glomus versiforme
A kind of Glomus mutans, species-specific technology, applied in the field of primer pairs for detection of Glomus mutans
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
specific Embodiment approach 1
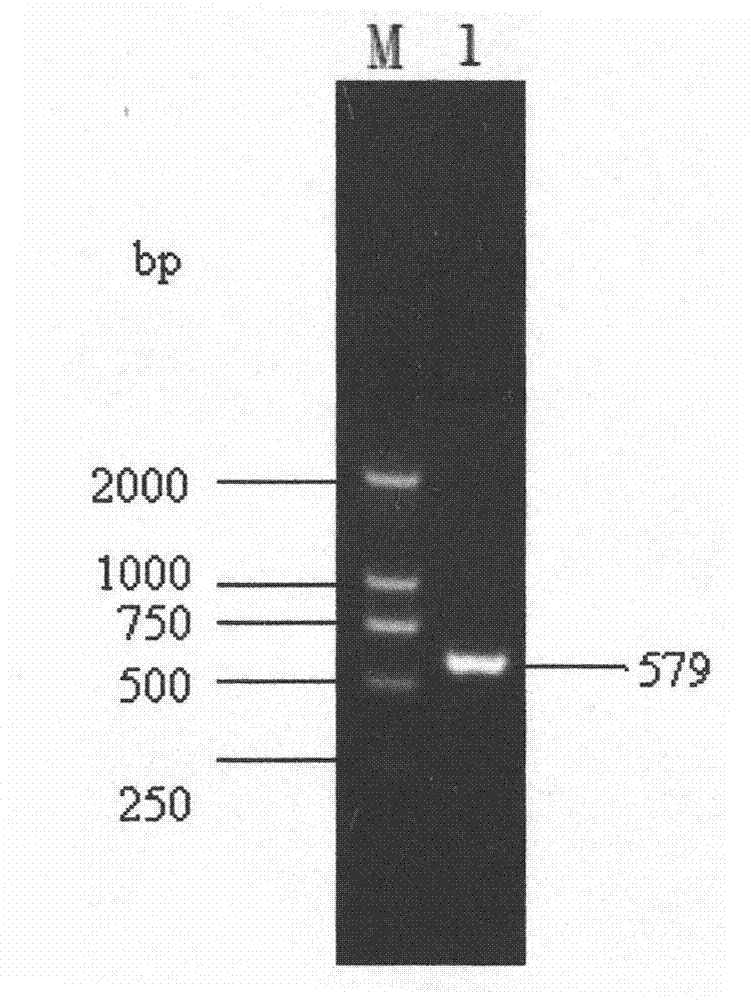
[0010] Specific embodiment 1: The species-specific primer pair used to detect Glomus mutans in this embodiment is named G.ver F / G.verR, and consists of a forward primer G.ver F and a reverse primer G.ver R, The base sequence of the forward primer G.ver F is 5'-GCATCTCCCAGGGTCTATAACA-3', and the base sequence of the reverse primer G.verR is 5'-GAGTGAAGAGGGAAAAGCTCAA-3'.
[0011] In this embodiment, the PCR amplification system for detecting the species-specific primer pair of Glomus mutans is 50 μL, consisting of 1.0 μL of template DNA, 4.0 μL of dNTP with a concentration of 2.5 mmol / L, and 5.0 μL of 10× buffer solution (containing Mg 2+ ), 5.0 μL of primers with a concentration of 10 pmol / L (final concentration of G.ver F and G.ver R), 0.2 μL of Taq DNA polymerase with a concentration of 5 U / μL, and the remaining sterile double-distilled water.
[0012] The PCR amplification reaction conditions of the species-specific primer pair used to detect Glomus mutans in this embodiment...
specific Embodiment approach 2
[0014] Embodiment 2: In this embodiment, the existing primer pair LR1 / G.ver is used for PCR amplification of Glomus mutans. The base sequence of the forward primer LR1 of the primer pair LR1 / G.ver is 5′-GCATATCAATAAGCGGAGGA-3′, and the base sequence of the reverse primer G.ver of the primer pair LR1 / G.ver is 5′-GTCTATAACACTCTCCCGAAG-3 '.
[0015] In this embodiment, the PCR amplification system for detecting the species-specific primer pair of Glomus mutans is 50 μL, consisting of 1.0 μL of template DNA, 4.0 μL of dNTP with a concentration of 2.5 mmol / L, and 5.0 μL of 10× buffer solution (containing Mg 2+ ), 5.0 μL of primers with a concentration of 10 pmol / L (the final concentration of LR1 and G.ver), 0.2 μL of Taq DNA polymerase with a concentration of 5 U / μL, and the remaining sterile double-distilled water.
[0016] PCR amplification reaction conditions: pre-denaturation at 95°C for 3min, denaturation at 93°C for 1min, annealing at 58°C for 1min, extension at 72°C for 1mi...
specific Embodiment approach 3
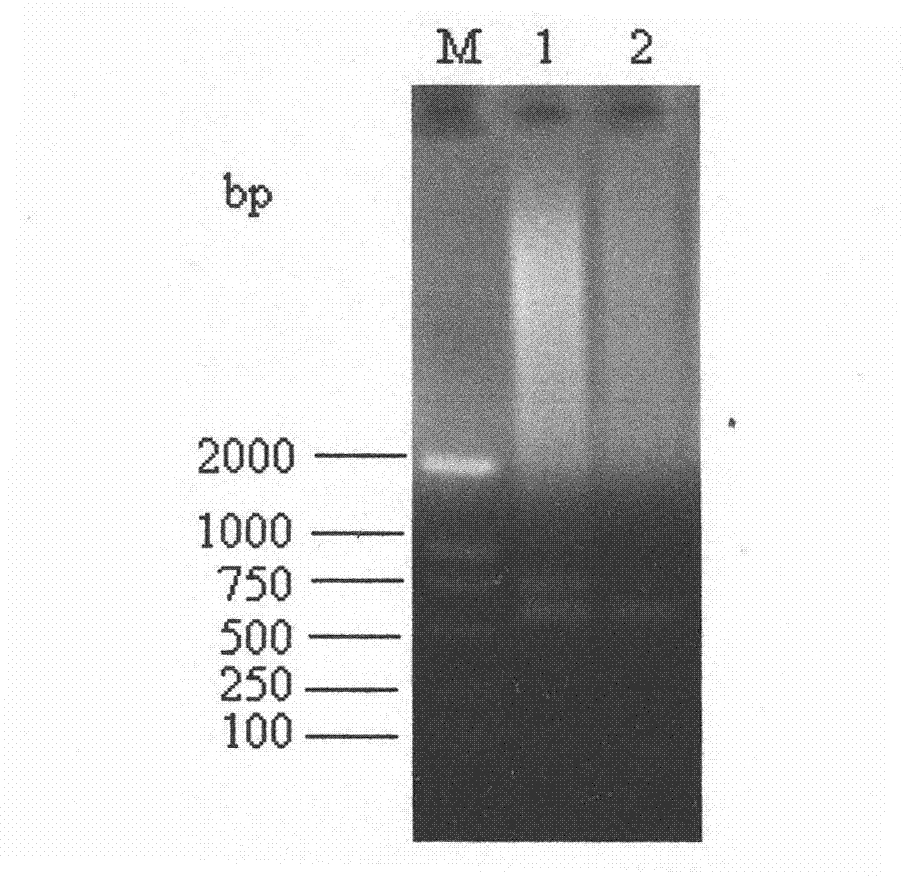
[0018] Specific embodiment three: In this embodiment, Glomus proteus is artificially added to the soil sample containing various AM fungi, and then the primer pair LR1 / G.ver and the primer pair G.ver F / G.ver R are used to carry out PCR amplification.
[0019] The PCR amplification of the primer pair G.ver F / G.ver R adopts the method of Embodiment 1, and the PCR amplification of the primer pair LR1 / G.ver adopts the method of Embodiment 2.
[0020] The soil samples containing multiple AM fungi in this embodiment were taken from the forest farm of Northeast Forestry University in September 2011.
[0021] The tested soil samples contained at least three kinds of AM fungi, Glomus intraradices, Glomus mosseae and Scutellospora calospora.
[0022] After PCR amplification, agarose gel electrophoresis was carried out for detection, and the detection results were as follows: image 3 shown. image 3 The middle primer pair LR1 / G.ver amplified two bands; while the primer pair G.ver F...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com