Applications of TRAIL truncated mutant in activating NF-kappaB and in inflammatory responses
A truncated and variant technology, applied to medical preparations containing active ingredients, anti-inflammatory agents, non-central analgesics, etc., can solve the problem of reduced ability of BX439 to activate NF-κB
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
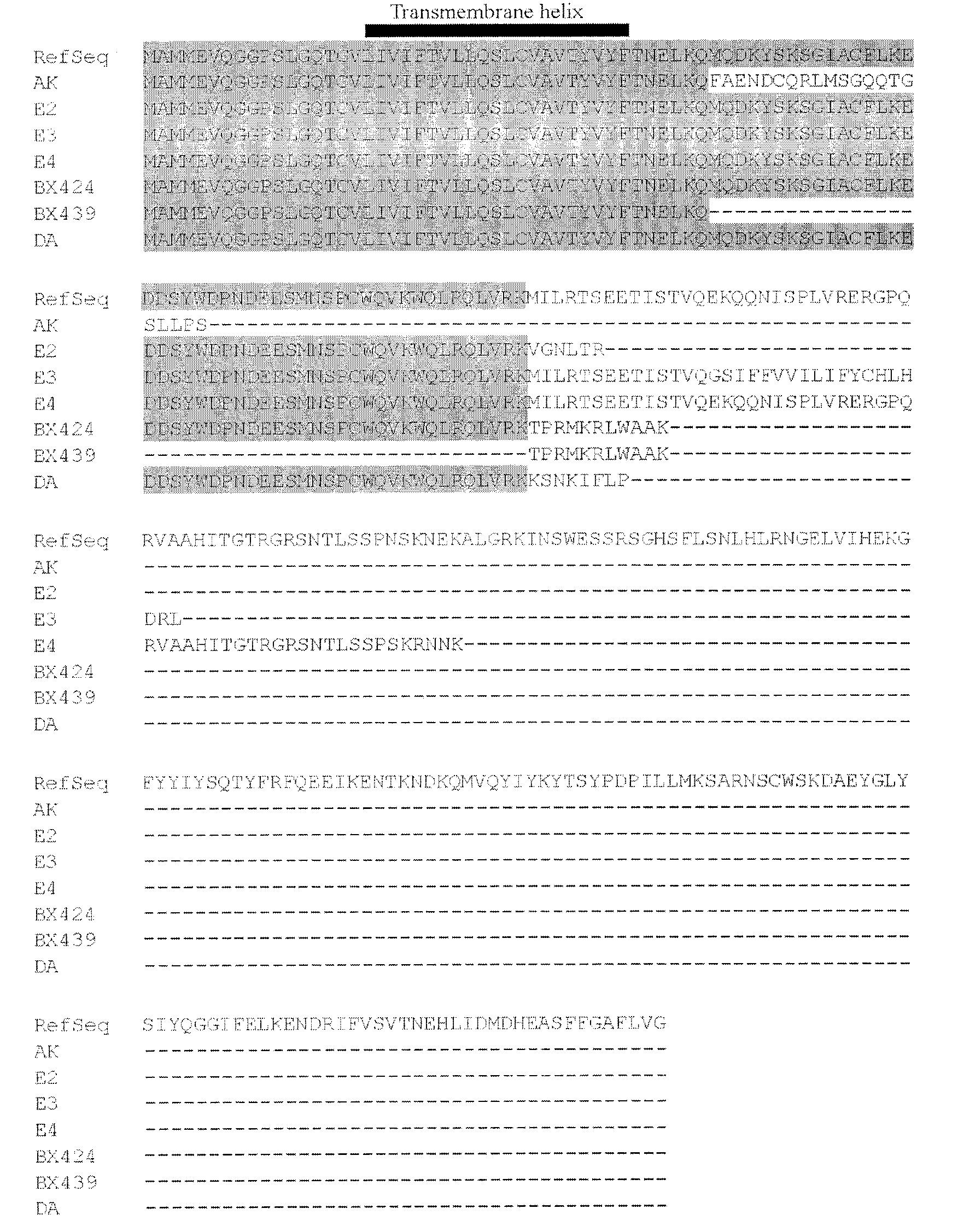
[0027] Structural analysis and protein sequence alignment of embodiment 1, 7 truncated TRAIL variants
[0028] Using EST database analysis, combined with UCSC genome browser viewing, we mapped the gene structure of 7 truncated TRAIL variants. The protein sequences of the seven truncated TRAIL variants were compared with those of RefSeq using bioinformatics software such as DNAstar and DNAMAN.
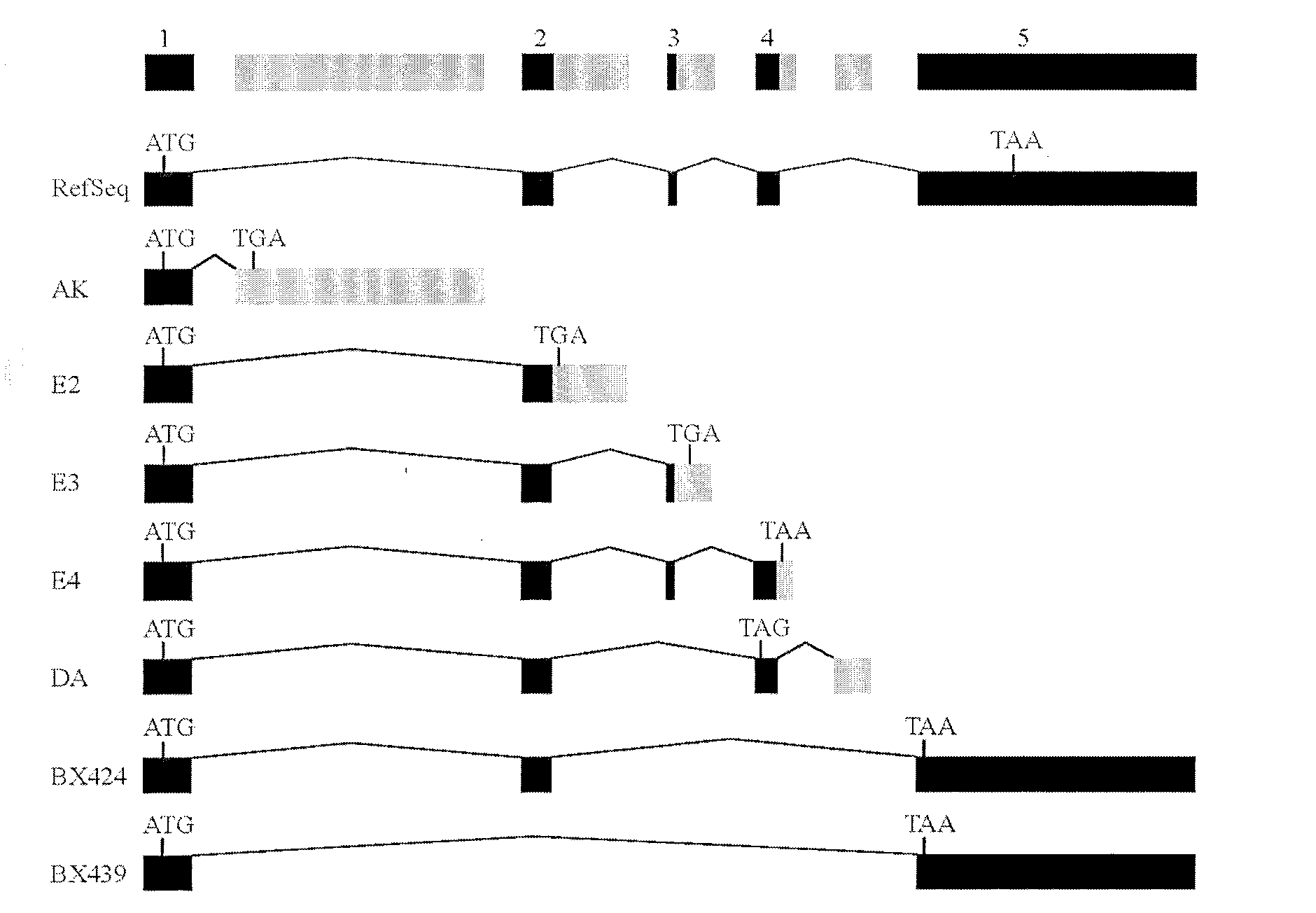
[0029] The results are shown in Figure 1, in which 1A shows that the intron boundaries spanned by all these variant sequences conform to the GT / AG rule, where AK (encoding 65 amino acids) and DA (encoding 98 amino acids) are in the first and DA lacks exon 2, and contains a new exon between exons 4 and 5. E2 (coding 88 amino acids), E3 (coding 123 amino acids) and E4 (coding 145 amino acids) were extended on the original exons 2, 3, and 4, respectively. BX424 (encoding 101 amino acids) lacks exons 3 and 4. BX439 lacks exons 2, 3, and 4. Black squares represent known exons, while gray...
Embodiment 2
[0030] Example 2, Expression of 7 truncated TRAIL variants in different cell lines
[0031] Eight kinds of cells (Raji, Jurkat, THP-1, HCT116, BT-325, HepG2, PC12, 293T, all purchased from ATCC, USA) were routinely cultured in a six-well plate (NEST Biotecl, 703001) at 300,000 / well 24 hours. RNA was extracted according to the instructions of the Invitrogen reverse transcription kit, reverse-transcribed into cDNA, PCR amplification conditions were 94°C for 5 minutes, denaturation at 94°C for 30 s, annealing at 58°C for 30 s, extension at 72°C for 40 s, and 30 cycles.
[0032] The result is as figure 2 As shown, the mRNA levels of 7 truncated TRAIL variants in different cell lines were detected by RT-PCR. Except that DA was only detected in THP-1, other variants were expressed in various cell lines.
Embodiment 3
[0033] Example 3, the expression of 7 truncated TRAIL variants on the cell membrane
[0034] HEK293T cells (purchased from ATCC, USA) were routinely cultured in a six-well plate (NEST Biotecl, 703001) at 300,000 / well for 18 hours, and 3 μg pCMV-sport6 (negative control), 7 truncated TRAIL variants Plasmid (pCMV vector), RefSeq (pCMV vector, positive control) According to the instructions, the plasmid was transfected into HEK293T cells using VigoFect (Vigorous Company) cationic transfection reagent. After continuing to culture for 24 hours, collect the cell culture supernatant in a centrifuge tube, trypsinize the cells, transfer them to a centrifuge tube, centrifuge at 1,500rpm for 5 minutes, discard the supernatant, wash the cells twice with PBS, add 2% FBS FACS blocking solution (PBS containing 2% FBS), the cells to be tested were made into a single-cell suspension with a density of approximately 5×10 5 / ml. Subsequently, the anti-Myc antibody (purchased from MBL Company) ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com