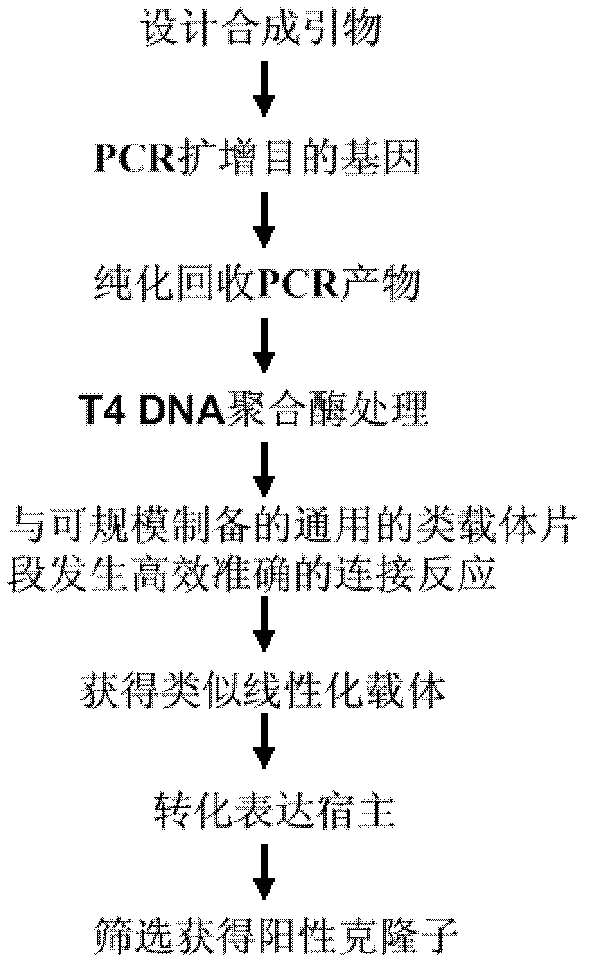
Construction method for linearized expression vector and simulated vector segments used for construction of linearized expression vector
A technology of expression vector and construction method, which can be applied in the direction of using vector to introduce foreign genetic material, recombinant DNA technology, etc., and can solve problems such as failure to be further popularized and applied.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
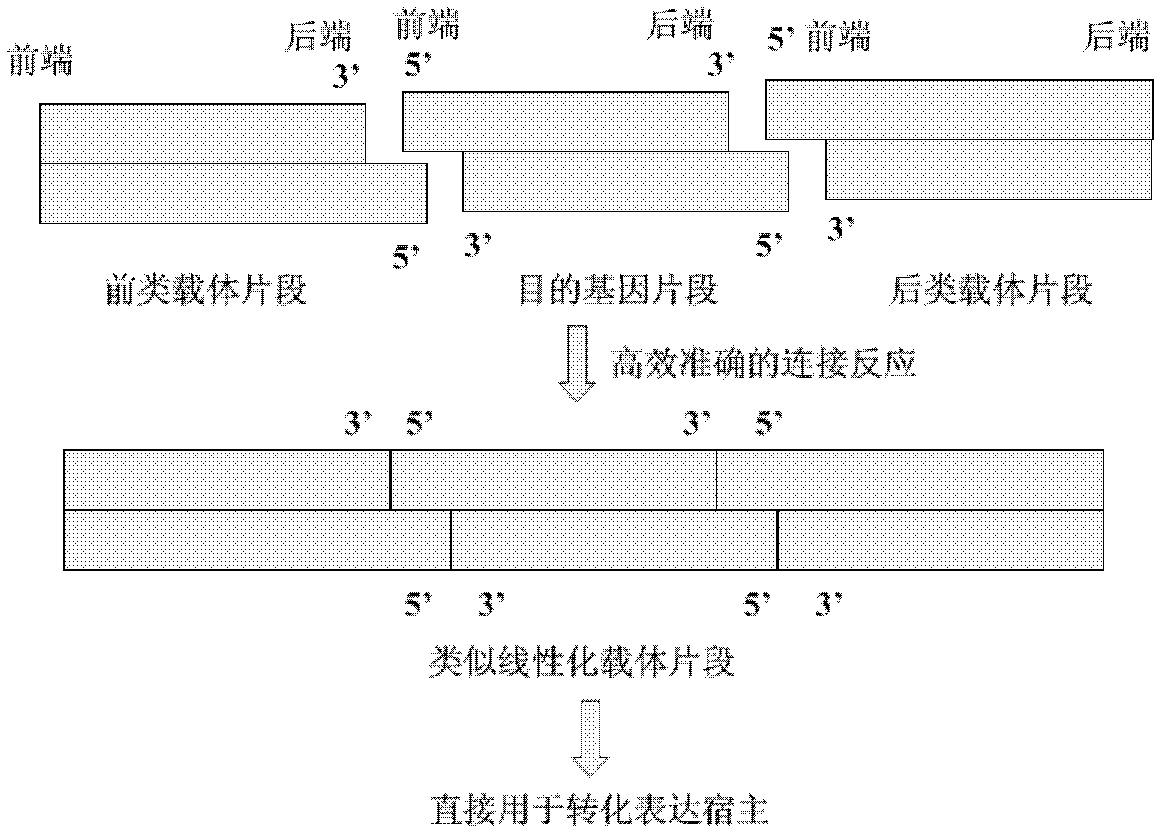
[0079] Preparing pre and post class vector fragments
[0080] In order to facilitate the preparation of front and back vector fragments, in the embodiment of the present invention, it is first constructed on a plasmid vector, and the spare front and back vector fragments can be obtained through the steps of plasmid purification, enzyme digestion and recovery.
[0081] The sequence SEQ ID NO: 1 containing the former vector fragment was constructed in plasmid pUC18 as follows: 1) with primers (5' ATGCGGATCCTACGTAAACATCCAAAGACGAAAGG 3' (SEQ ID NO: 3), 5' ATGCCTGCAGGTAGACTCGATCCTTCGAATAATTAG 3' (SEQ ID NO: 4)) Amplify the sequence of about 7bp-944bp of the plasmid pPIC9K, recover the amplified product from the gel and double-digest it with restriction endonucleases BamH I and Pst I, and recover the digested product from the gel for later use; Endonucleases BamH I and Pst I were double-digested and gel-recovered for later use; 3) Ligated the enzyme-digested vector and the fragment ...
Embodiment 2
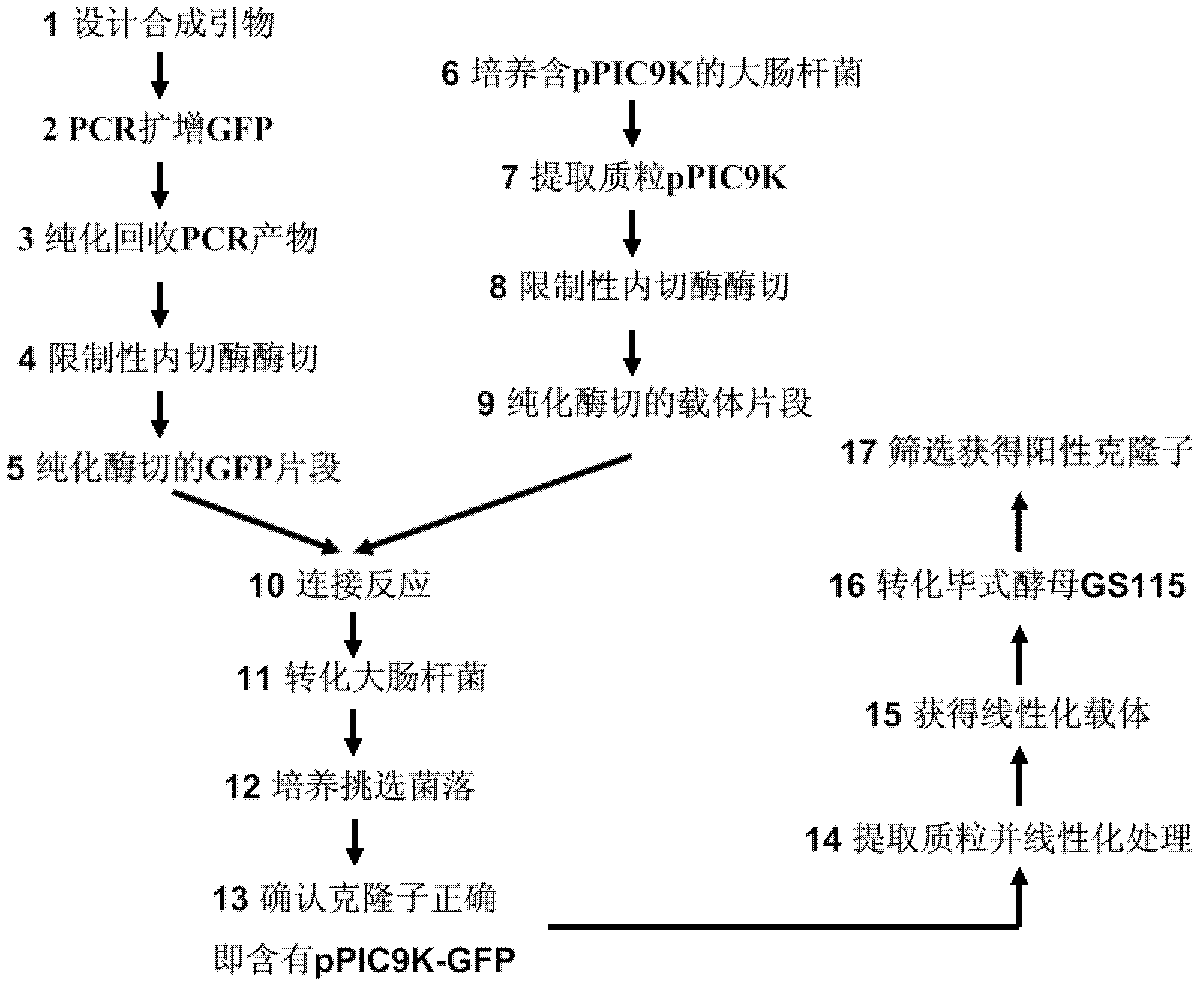
[0091] Preparation of target gene fragments
[0092] Design two primers to amplify the green fluorescent protein GFP gene, and add the base CTA to the 5' end of the upstream primer (note: because the first amino acid is coded by ATG, in order to ensure the correct reading frame, the first base of the GFP gene A is replaced by A in the CTA base), the base GTCA and stop codon are added to the 5' end of the downstream primer, and the sequence of the primer is as follows:
[0093] Upstream primer 5' CTAs TGGTGAGCAAGG 3' (SEQ ID NO: 9)
[0094] downstream primer 5' GTCA TTATACTTGTACAGC 3' (SEQ ID NO: 10)
[0095] The conditions for PCR amplification were: 10×Pyrobest DNA Polymorase Buffer, 5 μL; dNTP (10 mM), 4 μL; upstream primer (20 μM), 1 μL; downstream primer (20 μM), 1 μL; plasmid pEGFP-C1 (GenBank Accession #: U55763), 1ng; Pyrobest DNA Polymerase (TaKaRa Code DR005A), 0.5-1U; add distilled water to a total volume of 50μL. The PCR cycle conditions are: 94°C for 5 min; ...
Embodiment 3
[0100] The above-prepared carrier-like fragments and target gene fragments were connected with E. coli ligase (E.coli DNA Ligase, TaKaRa Code D2160A), and the reaction system was: carrier-like fragments, 10 pmol each; target gene fragments, 10 pmol; 10×E. coli DNA Ligase Buffer, 10 μL; 10×BSA (0.05%) 10 μL; E.coli DNA Ligase, 3U; add distilled water to a total volume of 100 μL, react at 16°C for 3 hours, and monitor the reaction by 0.8-1% agarose gel electrophoresis (result Such as Figure 5 : Swimming lane 3 is the test result of ligation for 3 hours, and swimming lane 2 is the control group without ligase), and the DNA fragment of 7.3kb was collected by gel for use. This fragment is the linearized plasmid of "pre-type vector-GFP-back-type vector" vector fragment. From the electrophoresis results, it can be seen that the connection efficiency is very efficient and accurate, and there are basically few wrongly connected bands. Density scanning of the 5.65kb fragment found tha...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com