Gene application in inhibition and apoptosis of glioma cell
A glioma cell and gene technology, applied in the application field of gene inhibition and apoptosis in glioma cells, can solve problems such as inability to cure, large brain damage, and easy recurrence
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0018] Embodiment one: dcf1 Gene amplification and enzyme digestion identification of recombinant plasmid pEGFP-N2-DCF1
[0019] 1. Using the cDNA of HeLa cells as a template for PCR amplification, in dcf1 EcoRI and BamHI restriction sites were introduced at both ends of the primers respectively. The upstream primer is (5'-CGGAATTCATGGCGGCGCCGAAGGGGAG-3'),
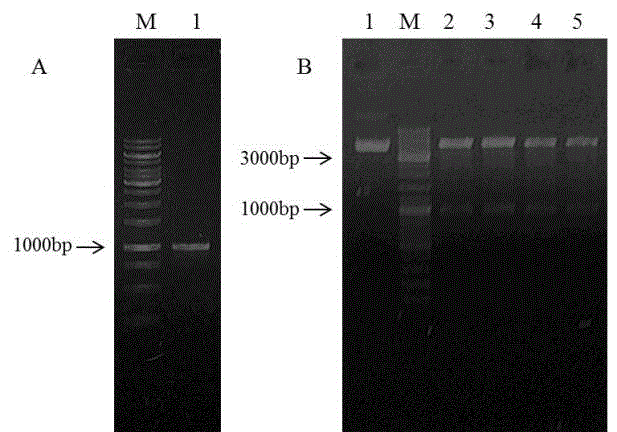
[0020] The downstream primer is (5'-CGGGATCCGTAAAATTTCAGAATGAGCA-3'), Tm is 53 degrees Celsius, and after 30 cycles, the target fragment of 972bp is obtained ( figure 1 A).
[0021]2. After the positive recombinant plasmid pEGFP-N2-DCF1 was obtained, the recombinant was identified by double enzyme digestion. Two enzymes, EcoRI and BamHI, were added to the enzyme digestion system at the same time. As a control, pEGFP-N2 EcoRI single enzyme digestion was used, and the reaction was performed at 37 degrees Celsius for 2 hours. The enzyme digestion results were identified by electrophoresis, and a band was released at 97...
Embodiment 2
[0022] Example 2: Transfection of plasmid pEGFP-N2-DCF1
[0023] After the U251 cell line was plated in a 24-well plate for 24 hours, the plasmid pEGFP-N2-DCF1 was transfected by Lipofectamine® 2000 Transfection Reagent cationic liposome transfection method. 1ul lipo2000 per well, 1ug plasmid pEGFP-
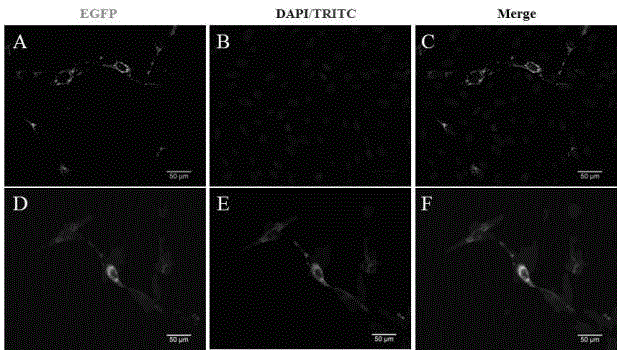
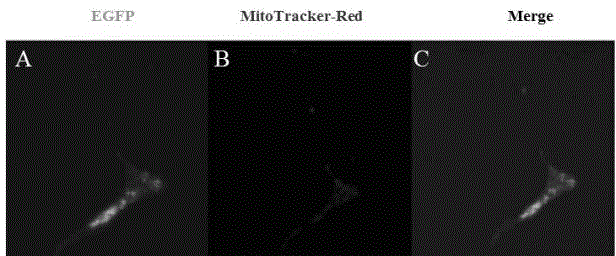
[0024] N2-DCF1 was dissolved in 50ul serum-free DMEM medium respectively, and after standing at room temperature for 5 minutes, the two were mixed evenly, and standing at room temperature for 20 minutes. The medium in the well plate was replaced with 400ul serum-free DMEM, and 100ul of the above mixed product was added. After culturing at 37°C for 6 hours, the medium was replaced with DMEM containing 10% fetal bovine serum. Fluorescence was detected 48 hours after transfection ( figure 2 A-C).
Embodiment 3
[0025] Example 3: Cell Immunofluorescence
[0026] Forty-eight hours after transfection of U251 cells, discard the medium and wash three times with PBS. Fix with 4% PFA for 10 minutes and wash with PBS three times. Permeabilize with 1‰Triton-X100 for 10 minutes, wash with PBS three times. Block with 2% BSA-PBS for 1 hour at room temperature. Dilute DCF1 rabbit polyclonal antibody (1:700) with 1% BSA-PBS, incubate at 37°C for 1 hour, wash with PBS three times. Dilute TRITC fluorescent secondary antibody (1:300) with 1% BSA-PBS, incubate at 37°C for 40 minutes in the dark, wash with PBS three times, and observe under a fluorescence microscope ( figure 2 D-F).
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com