Detection method of small non-messenger RNA
A non-coding and qualitative detection technology, applied in the field of non-coding small RNA detection, can solve the problems of increasing the difficulty of low-abundance non-coding small RNA detection, increasing the risk of small RNA degradation, and restricting promotion and application. Ease of follow-up detection, reduction of experimental costs, and wide application
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0036] step 1
[0037] Synthetic oligonucleotide sequence: 5'-ATTTGCGTGTCATCCTTGCG-3', this sequence is completely complementary to the U6 sequence, the probe is short, and it is easy to bind and pair with the complementary sequence.
[0038] Using terminal transferase (TdT) to label biotin at the 3' end of the oligonucleotide, the reaction system is as follows:
[0039] Ultrapure water 25μl
[0040] 5-fold dilution TdT reaction buffer 10μl
[0041] 5μl of oligonucleotide probe to be labeled
[0042] Biotin-11-UTP 5μl
[0043] TdT (2U / μl) 5μl
[0044] Reaction process: water bath at 37°C for 30min, then add 2.5μl EDTA (0.2M) to stop the reaction, add 50μl chloroform:isoamyl alcohol (24:1) mixed reagent, mix well and centrifuge (12000g, 2-3min), take the supernatant 40-50μl for storage.
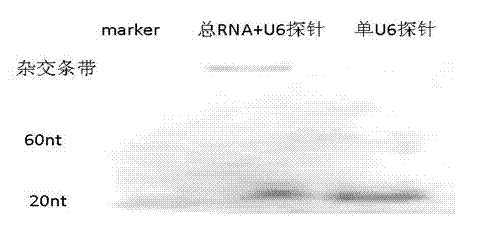
[0045] After the probe labeling is completed, the labeling effectiveness is tested: take different concentrations of labeled probe 1 μM, 2 μM, 5 μM, and 10 μM, add the loading buffer, ...
Embodiment 2
[0049] Referring to Example 1, different annealing conditions are used for liquid phase hybridization in this example.
[0050] Annealing condition 1: Set the temperature gradually on the PCR machine, 95 ° C for 2 min, and then drop 1 ° C every 90s until the temperature is 25 ° C, and maintain for a long time.
[0051] Annealing condition two: water bath at 95°C for 5min; then at 42°C for 2-3 hours.
[0052] refer to image 3 , the results show that the probe and target RNA can be hybridized to form a hybrid chain under two different liquid phase hybridization conditions. In terms of operation time, the action time of the PCR instrument is shorter than that of the water bath, the cooling process is mild, and the automatic control does not require human intervention; the action time of the water bath is prolonged, and the annealing is relatively mild. It is beautiful and clear, and non-specific binding is less than PCR instrument conditions. In short, both the water bath and...
Embodiment 3
[0054] Referring to Example 1, in this example, different hybridization buffers were used for liquid phase hybridization.
[0055] Hybridization buffer A: 30mmol / L phosphate buffer, 0.3mmol / L NaCl, 10mmol / LEDTA
[0056] Hybridization Buffer B: Tris-HCl (pH 8.5) 100mM, KCl 500nM, MgCl 2 15mM, 1% Tritox-100
[0057] Hybridization Buffer C: Tris-HCl (pH 7.5) 100 mM, 1 M NaCl, 10 mM EDTA
[0058] Hybridization buffer D: DEPC-treated H 2 O
[0059] Take 5 μg of fresh total RNA samples, 5 μl of U6 probe 4 groups, add equal volumes of different hybridization buffers, and place in a water bath at 95°C for 5 minutes; then at 42°C for 2-3 hours, fully hybridize, and then 15% non-denaturation Polyacrylamide gel electrophoresis detection.
[0060] refer to Figure 4 , the results show that under the same renaturation conditions, both phosphate buffer and Tris-HCl buffer can be used for liquid phase hybridization buffer, but the hybridization effect of phosphate buffer is obviously ...
PUM
 Login to view more
Login to view more Abstract
Description
Claims
Application Information
 Login to view more
Login to view more - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap