Synergistic effect of multiple bacterial strains on degradation of polyphenol pollutants and research method thereof
A synergistic, contaminant technology, applied in the directions of microorganism-based methods, biochemical equipment and methods, and microbial assay/inspection, which can solve problems such as refractory degradation, refractory microbial utilization, and few studies and reports
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
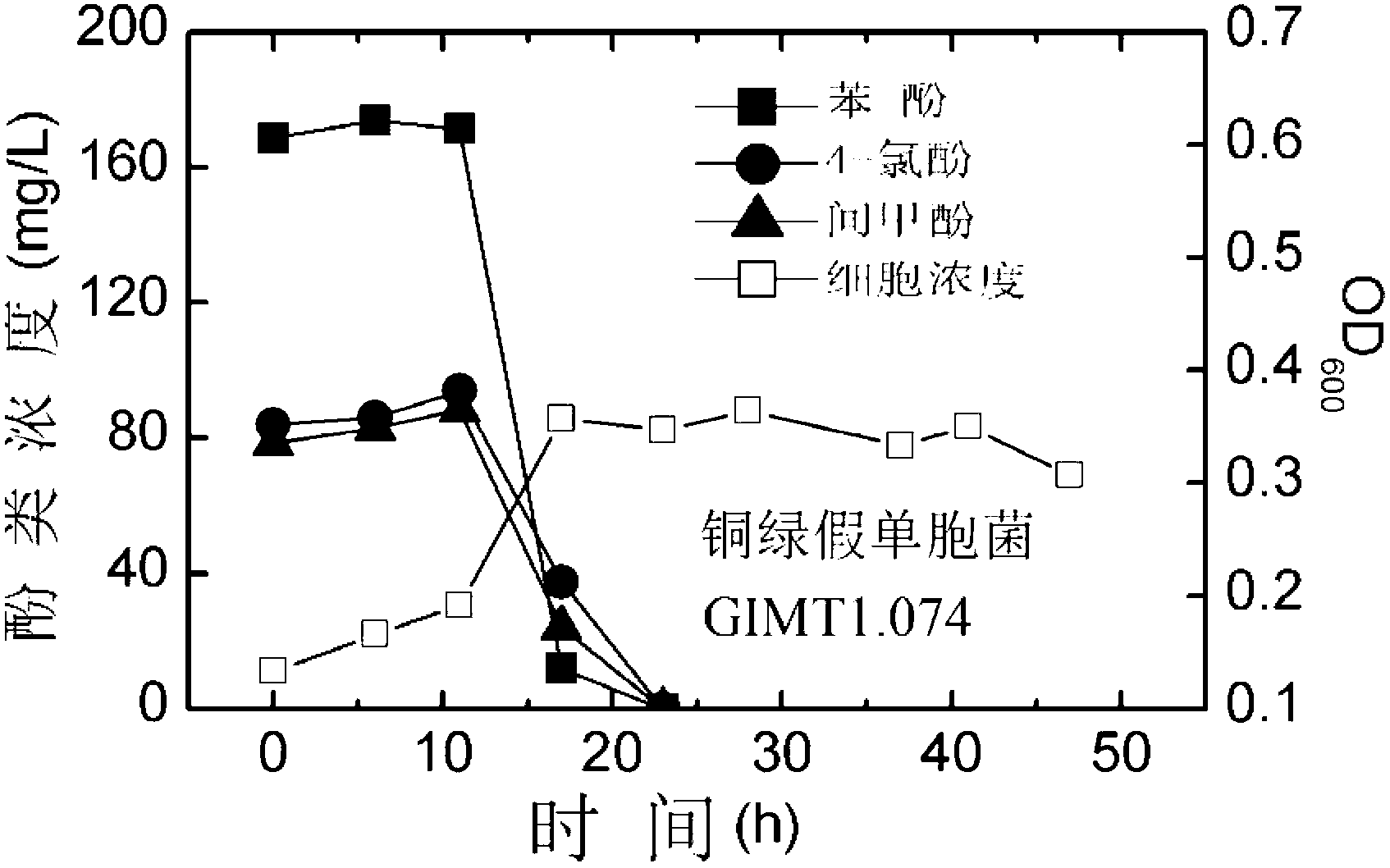
[0041] Degradation of polyphenol pollutants by Pseudomonas aeruginosa GIMT1.074
[0042] Such as figure 1 shown, according to the final OD 600 When the value is 0.1, Pseudomonas aeruginosa GIMT1.074 is inoculated in 50 mL of inorganic salt culture solution containing initial phenol concentration of 200 mg / L, initial 4-chlorophenol concentration of 100 mg / L and initial m-cresol concentration of 100 mg / L. 250mL Erlenmeyer flask. Place in a shaker at 30°C and 200rpm for cultivation. Take out 5 mL of culture medium every 6 hours, measure the concentration of cells in the culture medium and the concentrations of the three remaining substrates. The phenol concentrations at 0h, 6h, 12h, 18h, and 24h were 168.58mg / L, 173.96mg / L, 171.39mg / L, 11.88mg / L, and 0mg / L; the 4-chlorophenol concentrations were 84.03mg / L, respectively. L, 86.02mg / L, 94.01mg / L, 37.57mg / L and 0mg / L; the concentrations of m-cresol were 78.48mg / L, 82.93mg / L, 88.43mg / L, 24.06mg / L and 0mg / L respectively L; cell v...
Embodiment 2
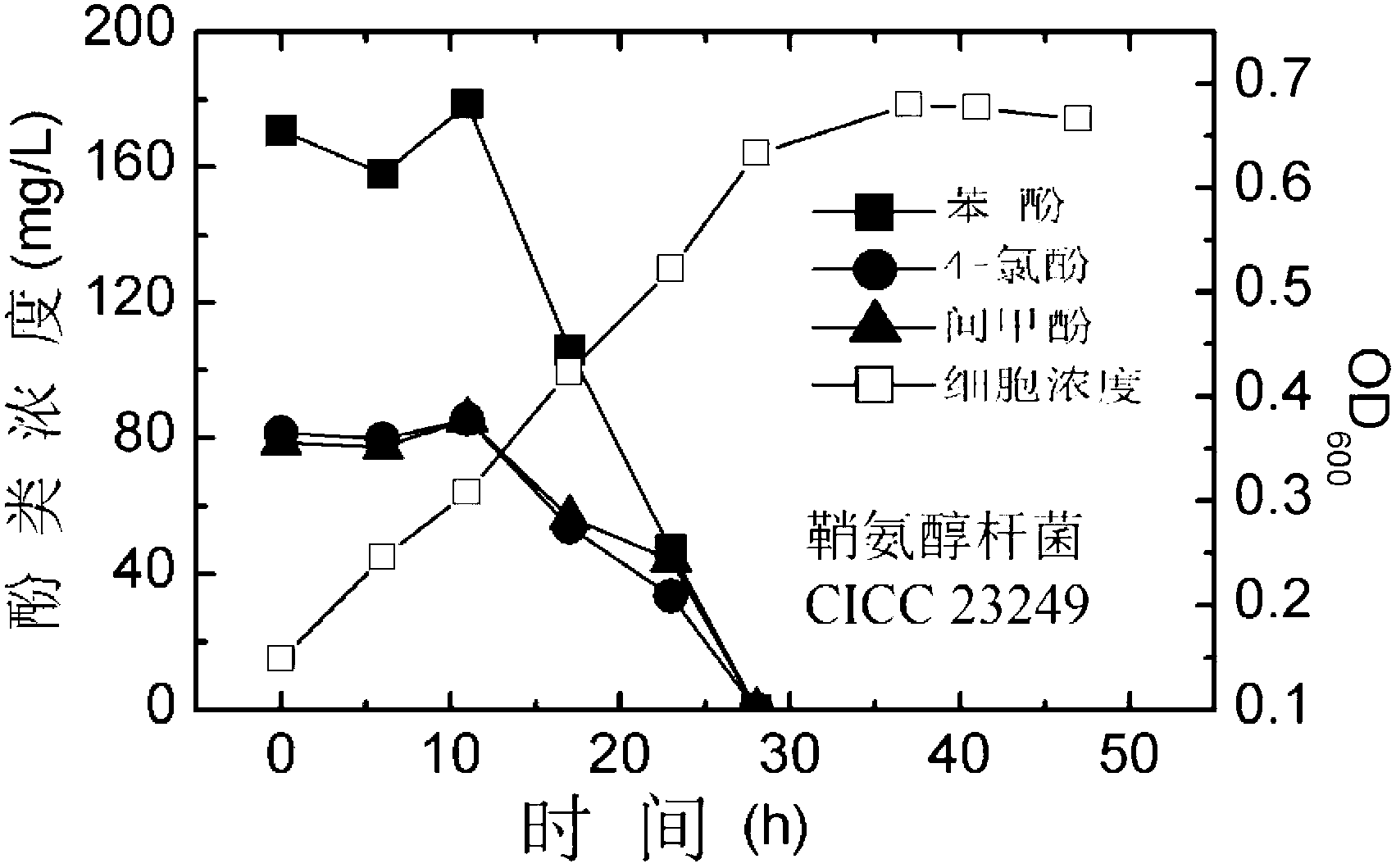
[0044] Degradation of Polyphenol Contaminants by Sphingobacterium CICC 23249
[0045] Such as figure 2 shown, according to the final OD 600 When the value is 0.1, Sphingobacterium CICC 23249 was inoculated in 250 mL of triangular triangle containing 50 mL of inorganic salt culture solution with initial phenol concentration of 200 mg / L, initial 4-chlorophenol concentration of 100 mg / L and initial m-cresol concentration of 100 mg / L. in the bottle. Place in a shaker at 30°C and 200rpm for cultivation. Take out 5 mL of culture medium every 6 hours, measure the concentration of cells in the culture medium and the concentrations of the three remaining substrates. The phenol concentrations at 0h, 6h, 12h, 18h, 24h and 30h were 170.85mg / L, 158.02mg / L, 178.71mg / L, 106.22mg / L, 47.11mg / L and 0mg / L, respectively; 4- Chlorophenol concentrations were 81.86mg / L, 79.65mg / L, 85.44mg / L, 53.84mg / L, 33.46mg / L and 0mg / L; m-cresol concentrations were 78.86mg / L, 77.34mg / L , 85.57mg / L, 56.47mg / ...
Embodiment 3
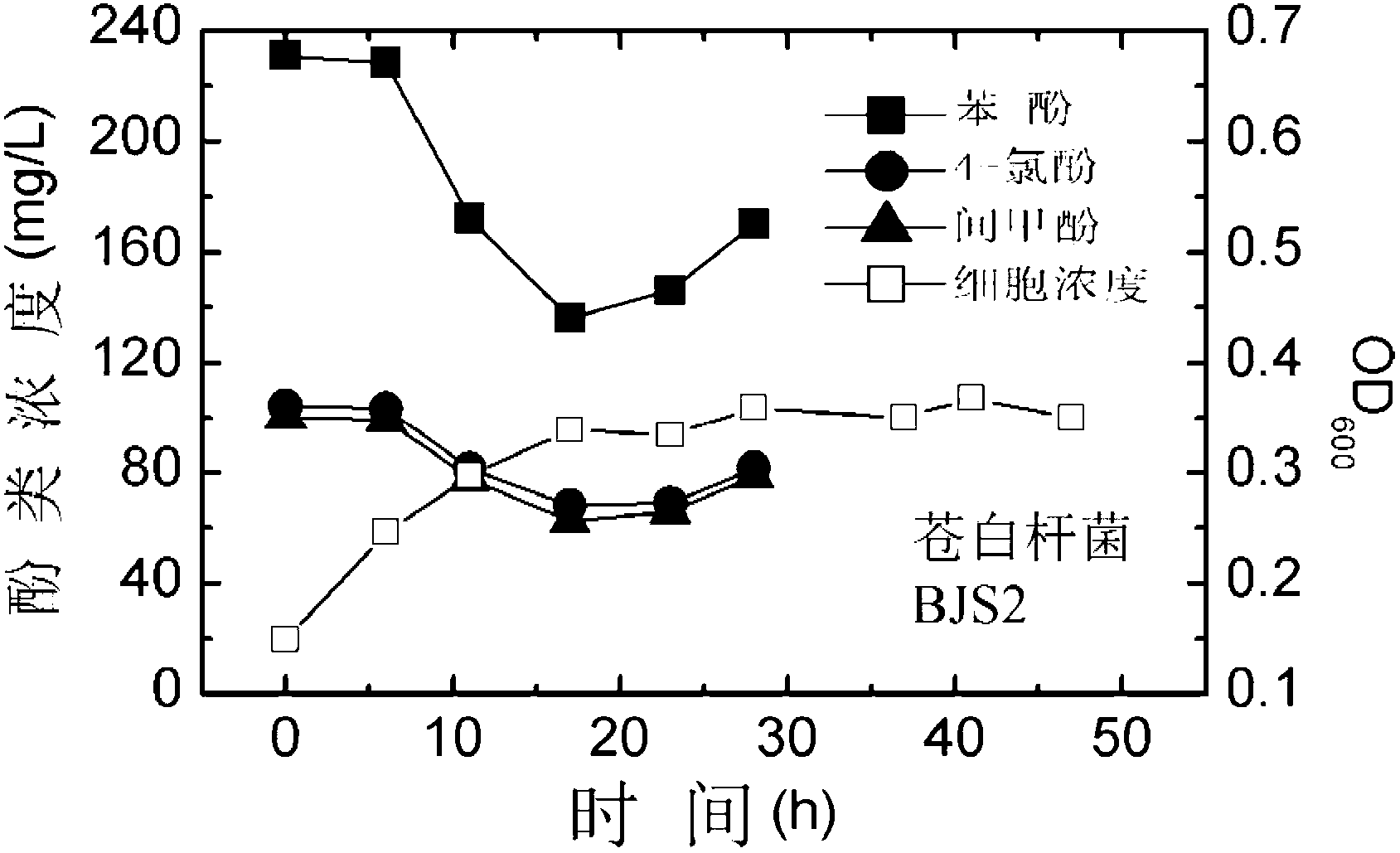
[0047] Degradation of Polyphenol Pollutants Using Pallidum pallidum BJS2
[0048] Such as image 3 shown, according to the final OD 600 The value is 0.1. The Pallidum pallidus BJS2 was inoculated in a 250mL Erlenmeyer flask containing 50mL inorganic salt culture solution with an initial phenol concentration of 200mg / L, an initial 4-chlorophenol concentration of 100mg / L and an initial m-cresol concentration of 100mg / L. Place in a shaker at 30°C and 200rpm for cultivation. Take out 5 mL of culture medium every 6 hours, measure the concentration of cells in the culture medium and the concentrations of the three remaining substrates. The phenol concentrations at 0h, 6h, 12h, 18h, 24h and 30h were 231.19mg / L, 228.55mg / L, 172.34mg / L, 136.32mg / L, 146.31mg / L and 170.19mg / L, respectively;4 -Chlorophenol concentrations were 104.13mg / L, 103.52mg / L, 81.43mg / L, 67.99mg / L69.06mg / L and 78.76mg / L; m-cresol concentrations were 100.05mg / L, 99.22mg / L L, 77.78mg / L, 62.54mg / L, 65.24mg / L and 78...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com