Construction method and application of Pichia stipitis large-fragment DNA (deoxyribonucleic acid) genome library
A Pichia stipitis and genome library technology, applied in the field of bioengineering, can solve problems such as unknown gene sequences
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
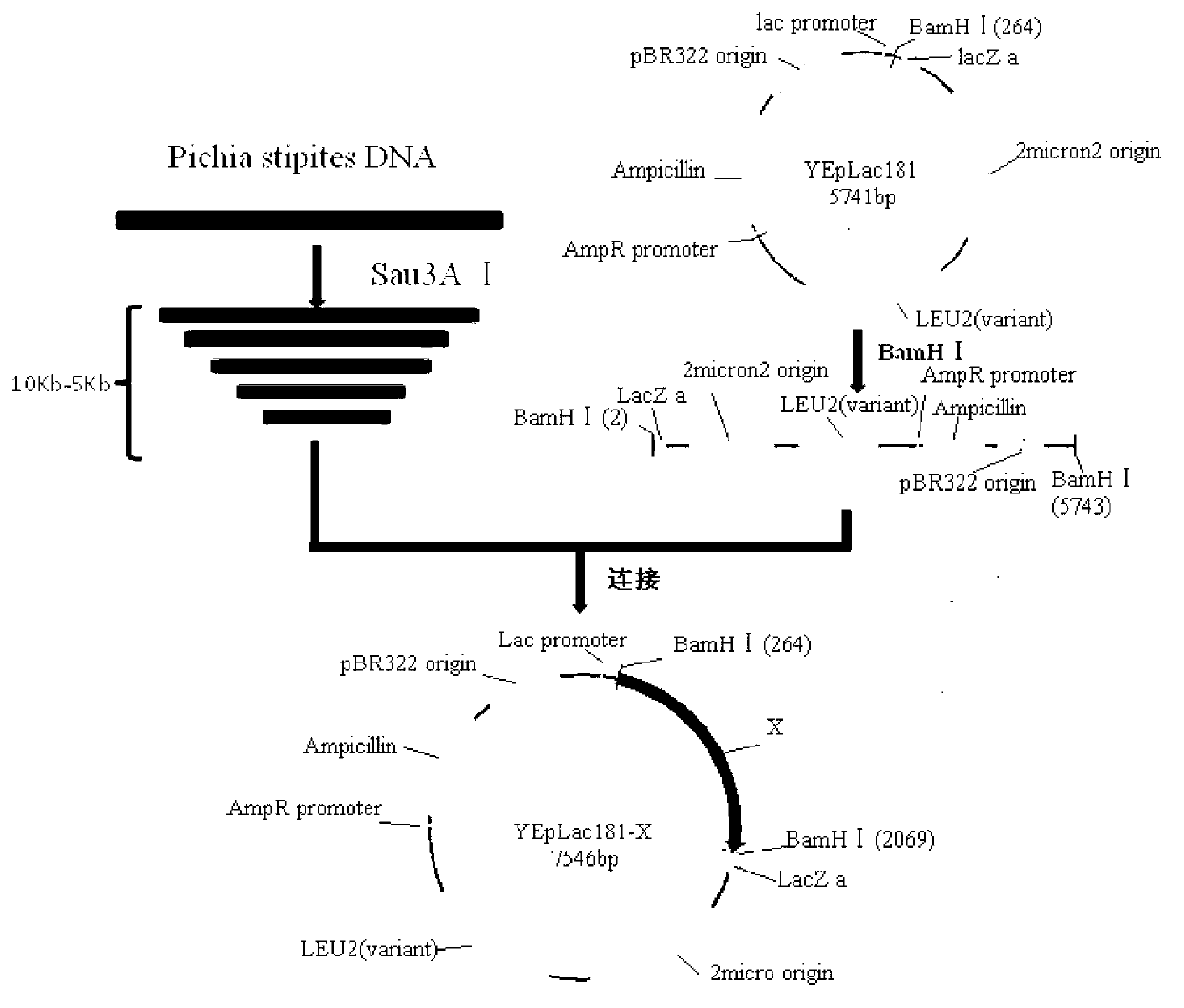
[0029] Example 1 Construction of Pichia stipitis CBS 5773 Large Fragment DNA Genomic Library
[0030] 1.1 Extraction of Pichia stipitis genomic DNA
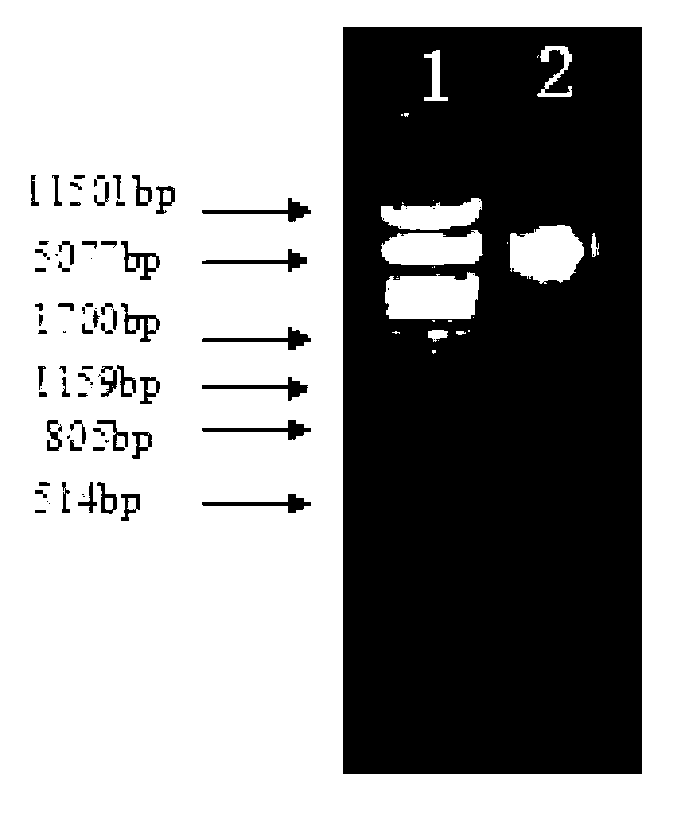
[0031] Take Pichia stipitis CBS 5773 and inoculate it in YEPD liquid medium, shake and cultivate overnight at 30°C, the shaker speed is 200r / min; take 2mL culture into a 5mL centrifuge tube, centrifuge at 6000r / min for 5min, discard Collect the supernatant, add 0.5mL Yeast Chromosome Extraction Reagent Ⅰ (0.1mol / L Na2EDTA-0.9mol / L Sorbitol, pH7.5), resuspend the bacteria; add 30μL 5mg / mL snail Enzyme, treat the bacterial cells at 37°C for 5 hours (at this time, most cells form protoplasts), centrifuge at 6000r / min for 5 minutes, discard the supernatant, and collect the bacterial cells; add 0.5mL yeast chromosome extraction reagent II (20mmol / L Na2EDTA-50mmol / L Tris-HCl, pH7.4), resuspend the cells, add 50μL of SDS with a mass volume concentration of 10%, and incubate at 65°C for 30min to lyse the cells; add 200μL of 5mol / L potas...
Embodiment 2
[0041] Example 2 Quality Evaluation of Pichia stipitis Large Fragment DNA Genomic Library
[0042] According to the number of positive clones obtained in the genomic library and the size of the average inserted fragment, according to the formula: library capacity = number of positive clones in the library × average size of inserted fragments / size of the biological genome, calculate the size of the genomic library The size of the storage capacity.
[0043] Among them, it is known that the genome size of Pichia stipitis CBS 5773 is 15441179bp, the number of positive clones in the library is 3000, and the average size of the inserted fragment is 5000bp. The calculated library capacity of the genome library is: library capacity = 3000×5000 / 15441179=97%, that is, the total length of the inserts contained in the library is 0.97 times the total length of the Pichiastipitis CBS 5773 genome, that is, any DNA sequence of Pichia stipitis was screened from the library The exact probabi...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com