Fluorescence detection method for trace tetracycline antibiotics
A tetracycline and fluorescence detection technology, which is applied in the direction of fluorescence/phosphorescence, material excitation analysis, etc., can solve the problems of complex operation and poor sensitivity, and achieve the effect of simple operation, high sensitivity and maintaining binding ability
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
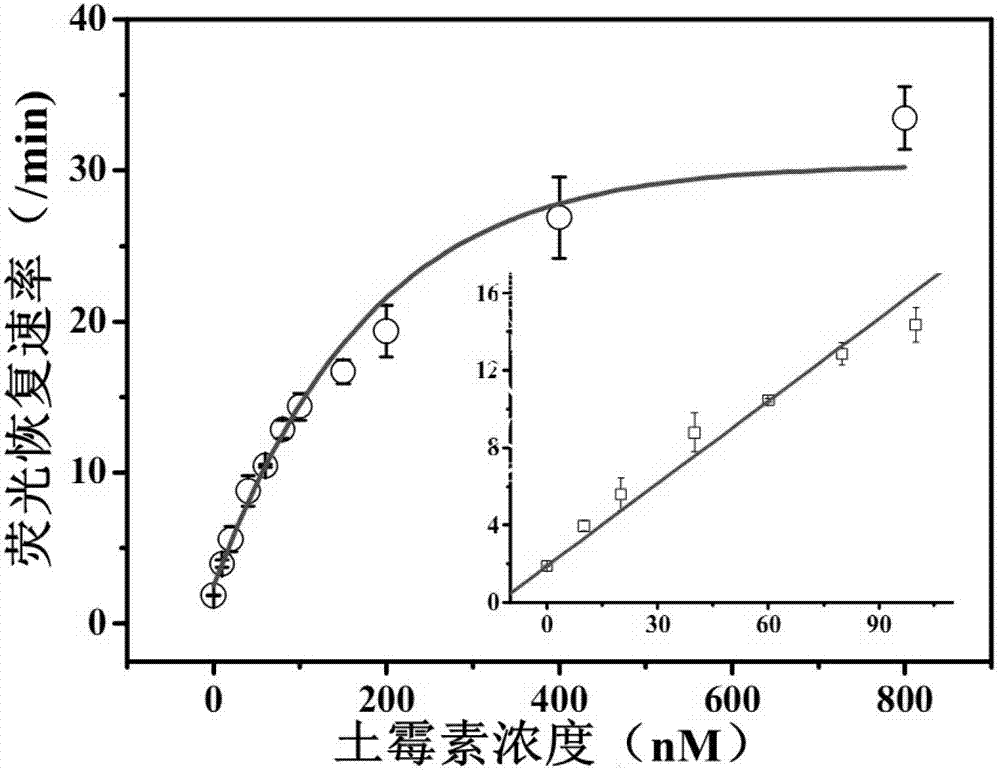
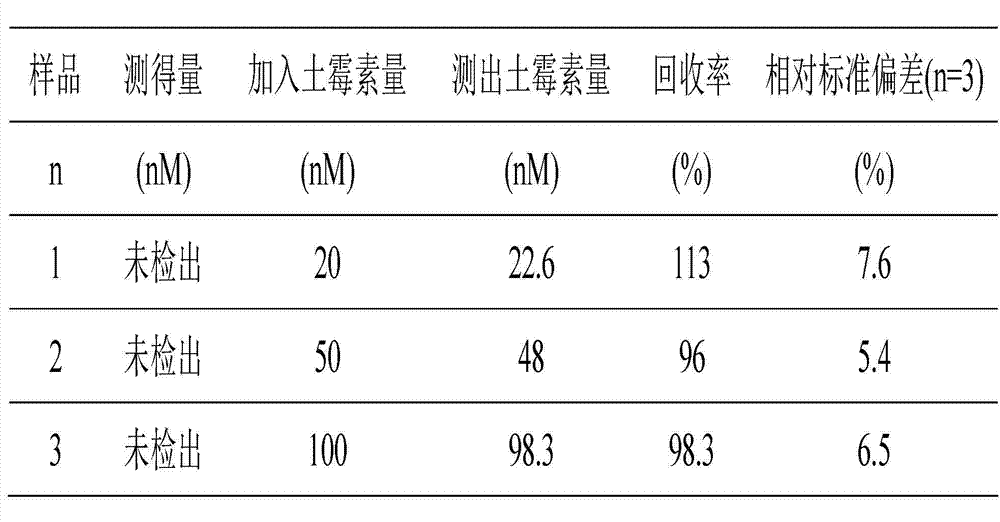
[0025] Example 1. Determination of Oxytetracycline Content in Milk Samples
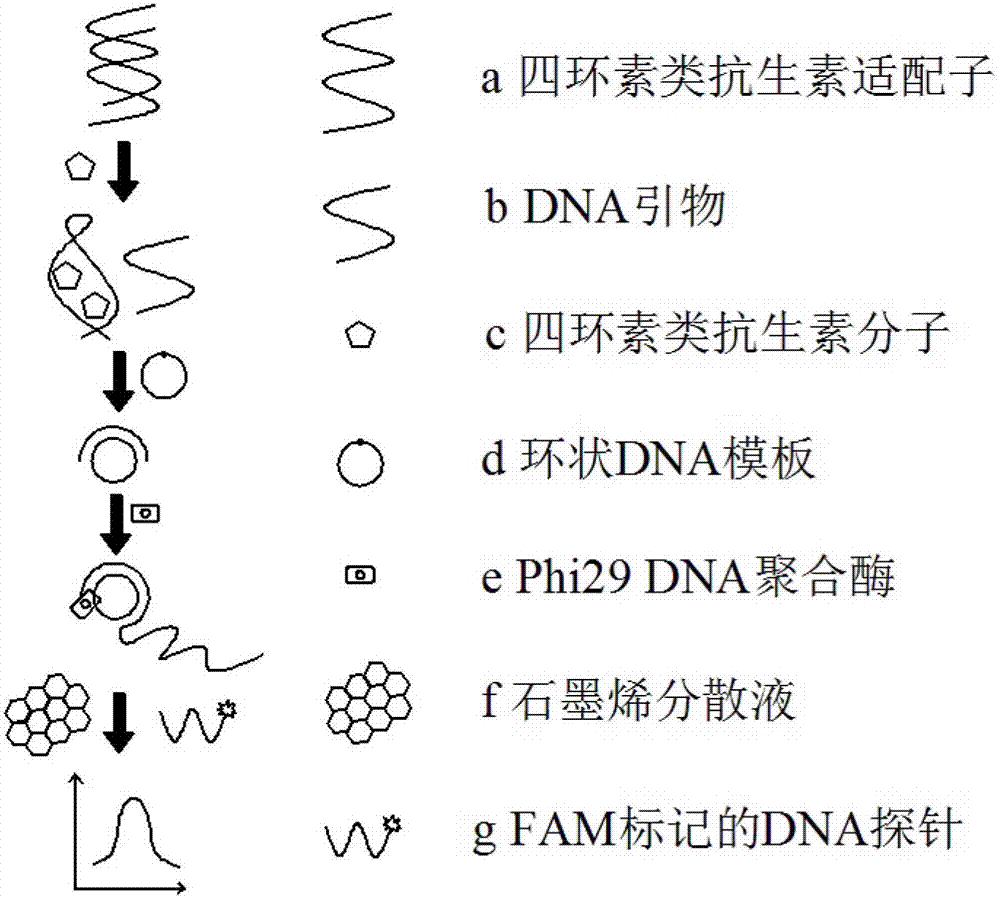
[0026] (1) Preparation of circular DNA template
[0027] Take 5 μL of phosphorylated padlock probe (final concentration 4 μM) and 10 μL DNA primer (final concentration 5.6 μM), mix well in 50 μL 1×T4 DNA ligase buffer, heat the water bath to 95 °C and keep 5 min, then annealing and cooling to 37 °C for 1 h, then adding T4 DNA ligase (final concentration 3.5 U / μL), 5 μL 0.05% BSA, and 50 μL high-purity water at 16 °C overnight. After the reaction is over, add 1 μL 10×Exonuclease I buffer, 1 μL 10×Exonuclease I buffer, Exonuclease I (final concentration 0.1 U / μL), and Exonuclease III (final concentration 0.1 U / μL) Incubate at 37 °C for 1 h to degrade unreacted single-stranded DNA primers. Heat the water bath to 90 °C and incubate for 10 min to terminate the reaction. Finally, the prepared circular DNA template was separated and purified with nanofiltration membrane (10 K, Pall, USA) to remove macromolecula...
Embodiment 2
[0043] Example 2. Determination of tetracycline antibiotics in lake water
[0044] (1) Preparation of circular DNA template
[0045] Take 5 μL of phosphorylated padlock probe (final concentration 2 μM) and 10 μL DNA primer (final concentration 4 μM) and mix well in 50 μL 1×T4 DNA ligase buffer, heat the water bath to 80 °C and keep 5 min, then annealing and cooling to 37 °C for 0.5 h, then adding T4 DNA ligase (final concentration 2 U / μL), 5 μL 0.05% BSA, and 50 μL high-purity water at 10 °C overnight. After the reaction, add 1 μL 10×Exonuclease I buffer, 1 μL 10×Exonuclease I buffer, Exonuclease I (final concentration 0.05 U / μL), and Exonuclease III (final concentration 0.05 U / μL) at 37 °C. Incubate for 0.5 h to degrade unreacted single-stranded DNA primers. Heat the water bath to 80 °C and incubate for 5 min to terminate the reaction. Finally, the prepared circular DNA template was separated and purified with nanofiltration membrane (10 K, Pall, USA) to remove macromolecular en...
Embodiment 3
[0061] Example 3 Determination of Oxytetracycline Content in Lake Water Sample
[0062] (1) Preparation of circular DNA template
[0063] Take 5 μL of phosphorylated padlock probe (final concentration 6 μM) and 10 μL DNA primer (final concentration 8 μM) and mix well in 50 μL 1×T4 DNA ligase buffer. Heat in a water bath to 95 °C and keep After 15 min, annealing and cooling to 37 °C for 2 h, then adding T4 DNA ligase (final concentration 6 U / μL), 5 μL 0.05% BSA, and 50 μL high-purity water at 25 °C overnight. After the reaction, add 1 μL 10×Exonuclease I buffer, 1 μL 10×Exonuclease I buffer, Exonuclease I (final concentration 0.2 U / μL), and Exonuclease III (final concentration 0.2 U / μL). Incubate at 37 °C for 2 h to degrade unreacted single-stranded DNA primers. Heat the water bath to 95 °C and incubate for 15 minutes to terminate the reaction. Finally, the prepared circular DNA template was separated and purified with nanofiltration membrane (10 K, Pall, USA) to remove macromolec...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com