Sorbose dehydrogenase and sorbosone dehydrogenase, and applications thereof
The technology of sorbose dehydrogenase and sorbone dehydrogenase is applied in the field of microbial biology and can solve the problems of long cycle, increased process control, nutrient composition of fermentation medium and energy waste, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0030] Example 1 Extraction of Gluconobacter oxydans CGMCC No.1.637 Genomic DNA
[0031] (1) Cultivate Gluconobacter oxidans CGMCC No.1.637 (purchased from the Institute of Microbiology, Chinese Academy of Sciences) until the OD value is about 2.0, and collect 5 mL of bacterial liquid by centrifugation at 4°C and 8000 rpm for 5 min.
[0032] (2) Add 450 μL TE, 50 μL 10% SDS, 5 μL proteinase K, and vortex at 1800 rpm to fully resuspend.
[0033] (3) Cleavage at 80°C for 10 minutes, let cool to room temperature, add 5 μL RNaseA, mix by inverting 10 times, and bathe in 37°C water for 2 hours.
[0034] (4) Add 30 μL of 3M KAC, shake at 1800 rpm for 30 seconds, and place at -20°C for 20 minutes.
[0035] (5) Centrifuge at 12000 rpm at 4°C for 20 min, and pipette the supernatant into another centrifuge tube.
[0036] (6) Add 60 μL of 5M NaCl and 300 μL of isopropanol, invert and mix 20 times, and place at -20°C for 20 minutes.
[0037] (7) Centrifuge at 12000 rpm and 4°C for 20 m...
Embodiment 3D
[0046] Embodiment 3DCIP and active electrophoresis detection activity
[0047] The pMD18T-SNDHSDH / DH5α cells were collected by centrifugation and lysed by ultrasound. After centrifugation, the supernatant was taken for activity detection by DCIP method and Native electrophoresis. Sorbose and xylose were used as substrates respectively, and the results showed SDH and SNDH activities (Table 2) .
[0048] Table 2 DCIP and activity electrophoresis detection activity results
[0049]
[0050] Note: "+" means no activity, "-" means no activity.
Embodiment 4
[0051] Example 4 Construction of pBBR1MCS2-SNDHSDH-1.637 plasmid
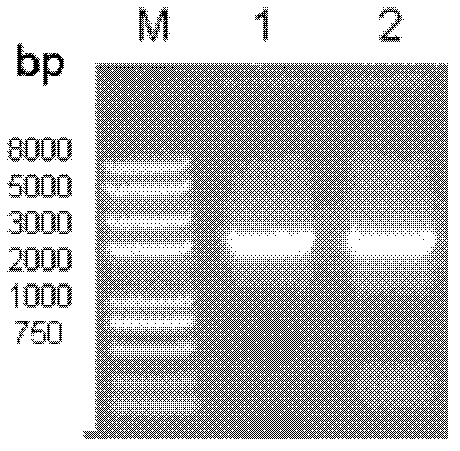
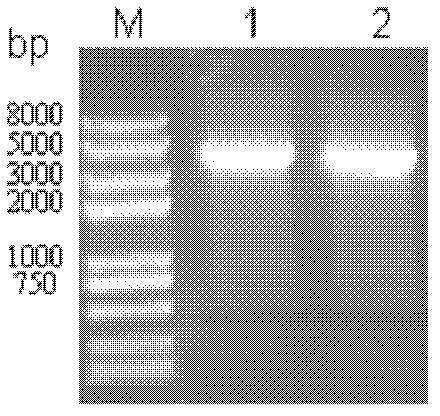
[0052] The successfully constructed pMD18T-SNDHSDH plasmid and pBBR1MCS2 (GenBank accession number U23751) plasmid vector were digested with Xba I / SalI respectively, the target fragment was recovered by the agarose gel recovery kit, ligated at 16°C (10 μL system) overnight, and transformed into DH5a , to screen for positive recombinants. Identification by bacterial liquid PCR ( Figure 6 ) and sequencing identification showed that the pBBR1MCS2-SNDHSDH plasmid was constructed successfully.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - Generate Ideas
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com