Crystallized oxalate decarboxylase and methods of use
A technology for oxalate decarboxylase and oxaluria, which is applied in the directions of biochemical equipment and methods, medical preparations containing active ingredients, enzymes, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
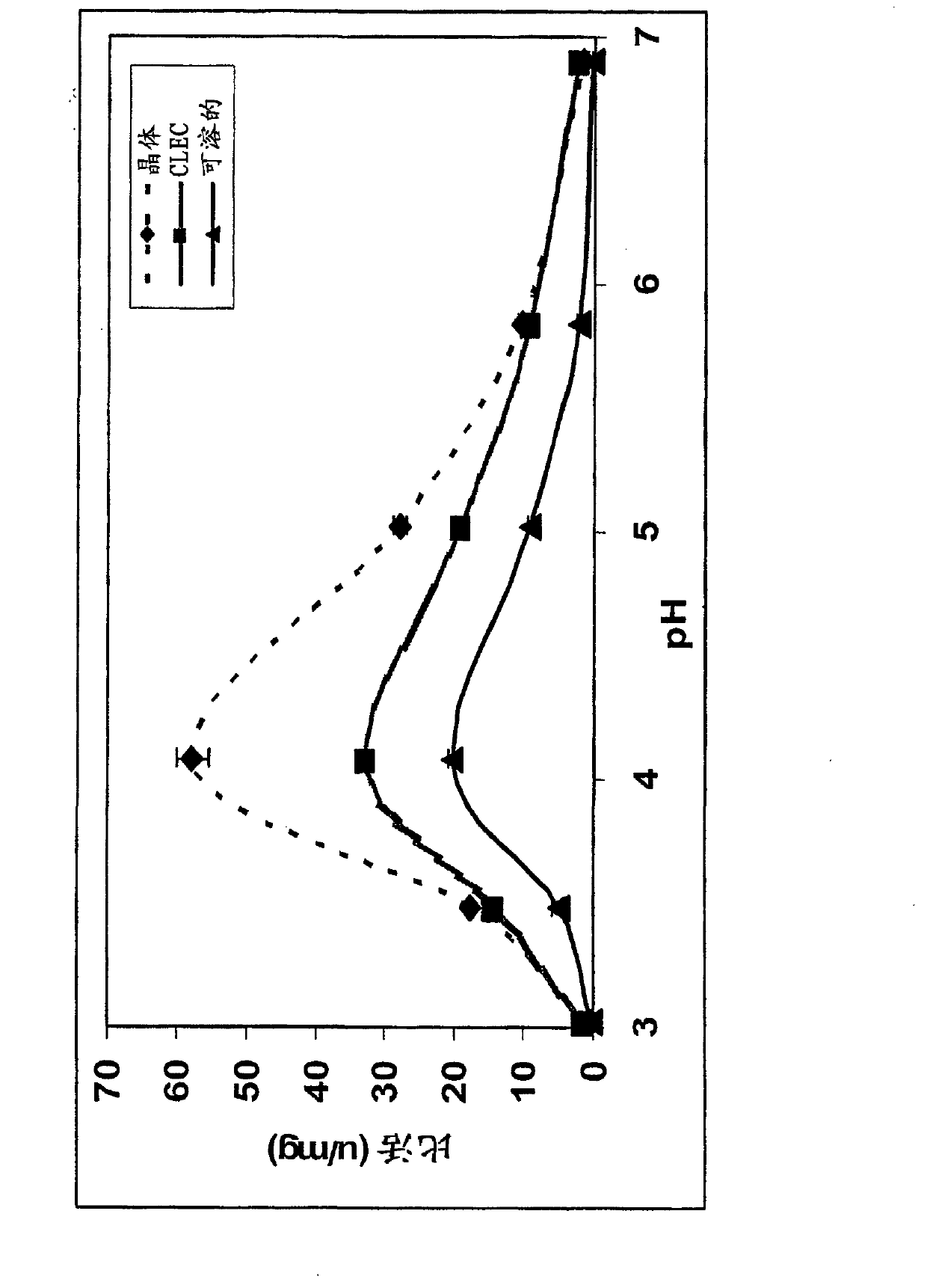
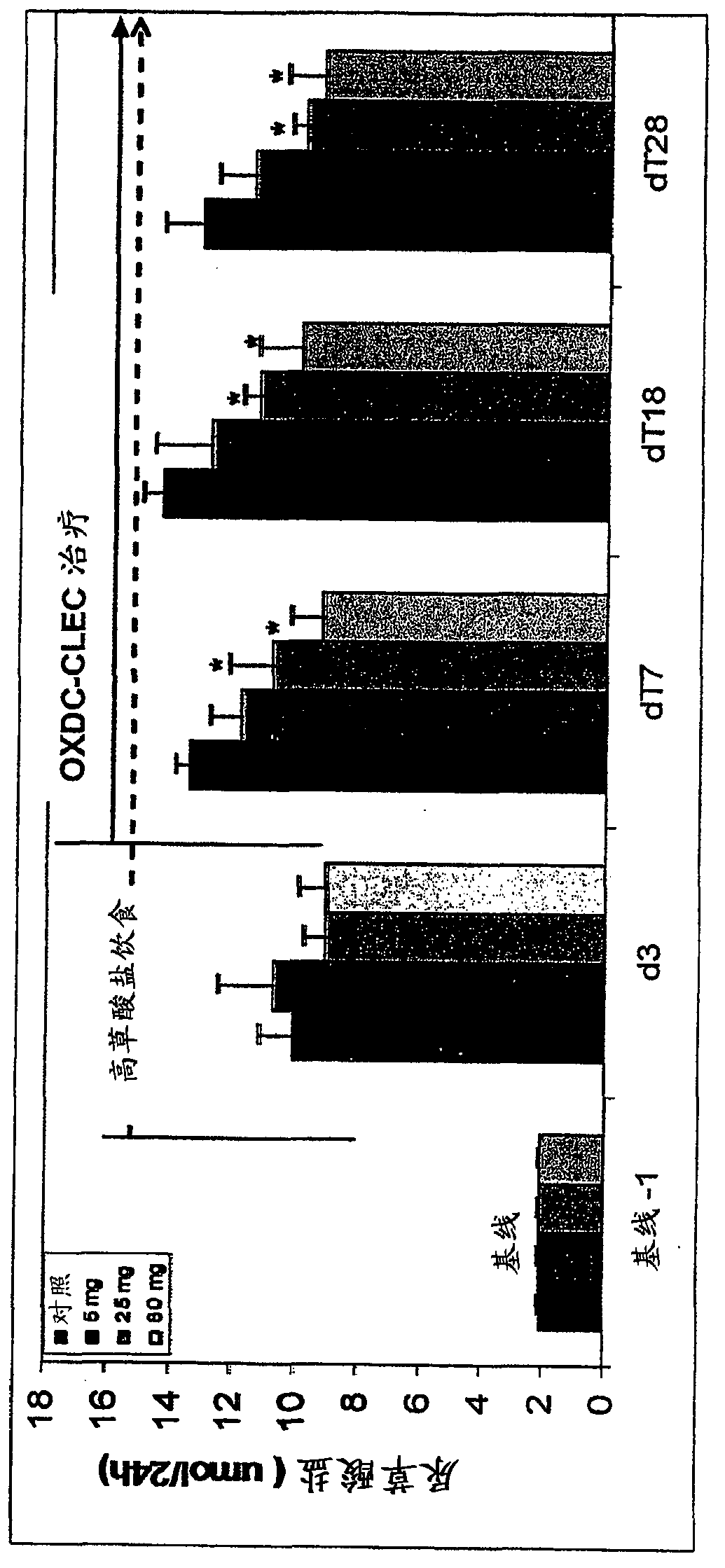
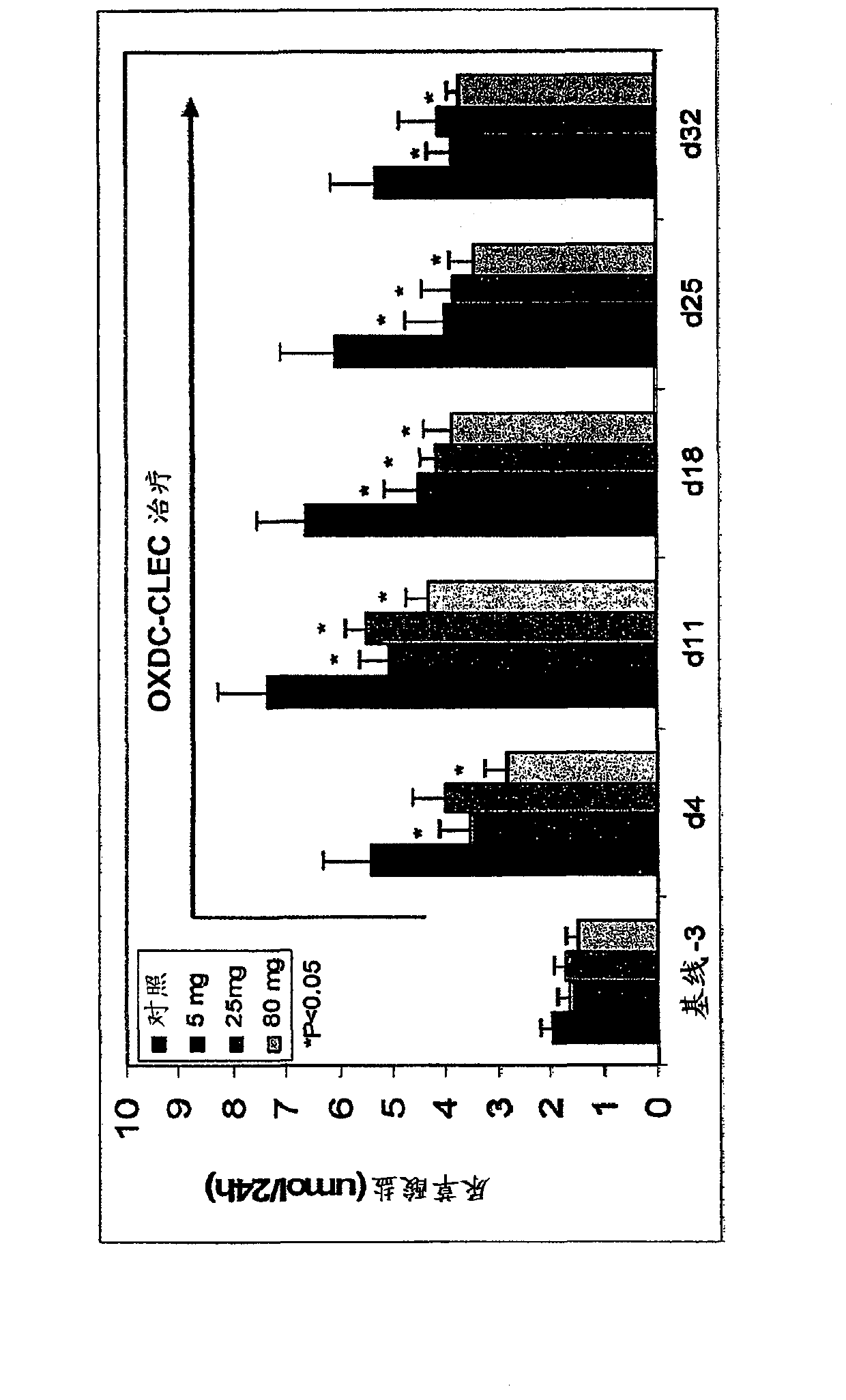
Image
Examples
Embodiment 1
[0135] Example 1. Fermentation and purification of oxalate decarboxylase.
[0136] Oxalate decarboxylase (OXDC) from Bacillus subtilis (B. subtilis) is a 261 kDa homohexameric protein composed of 6 identical monomers. Each monomer contains 385 amino acids with a calculated molecular weight of -43-44 kDa and an isoelectric point of 5.2. The OXDC gene formerly known as yvrK was PCR amplified using Bacillus subtilis genomic DNA as a template.
[0137] The amplified OXDC gene was first cloned into the pCRII vector (Invitrogen, Carlsbad, CA), then subcloned, and expressed from the pET-11a expression vector using E. coli BL21(DE3)pLysS cells. This gene expression in the pET-11a vector is under the control of the T7 promoter, induced by IPTG (isopropyl-β-D-thiogalactoside), which regulates OXDC expression.
[0138]Fermentation was used to achieve high level expression of recombinant OXDC in E. coli. In the presence of casein hydrolyzate (USB Corporation, Cleveland, OH) or soy pe...
Embodiment 2
[0142] Example 2. Using mild denaturant concentrations and subsequent anion exchange chromatography from solubilized Granular crystalline oxalate decarboxylase.
[0143] OXDC pellets refrigerated at -20°C were used to prepare OXDC crystals.
[0144] In this procedure, particles are dissolved under mild conditions of denaturant concentration and pH. The solubilized protein is then refolded using an anion exchange matrix column.
[0145] The particles were dissolved in 2M urea, 100 mM Tris pH 10.0, 10 mM DTT and 100 mM NaCl (1:10 w / v). The solution was stirred at room temperature (RT) for 2 h, then centrifuged at 15K for 30 min at 4°C. The supernatant was carefully decanted and stored. The pellets were carefully weighed and stored separately.
[0146] Under constant and gentle agitation, at a flow rate of 10 ml / min, the solubilized particles in the supernatant were added dropwise to 10 volumes of a solution consisting of 2M urea, 100 mM Tris pH 8.0, 1 mM DTT and 1 mM MnC...
Embodiment 3
[0150] Example 3. Using high denaturant concentrations and subsequent anion exchange chromatography, by dissolution The particles crystallize oxalate decarboxylase.
[0151] By this method, particles were dissolved in 5M urea, 5OmM Tris pH8.6, 10OmM NaCl, 1OmM DTT (1 :5 w / v). The solution was stirred at room temperature for 2 h and then centrifuged at 15K for 30 min at 4°C. The supernatant was carefully decanted and stored. The pellets were carefully weighed and stored separately.
[0152] Prepare anion exchange chromatography columns by packing Q Sepharose matrix into glass columns. The column was attached to the FPLC and equilibrated by washing with 3 column volumes (CV) of 4M urea, 100 mM Tris pH 8.6 and 10 mM DTT. With 7 column volumes of 100mM NaCl, 50mM Tris pH8, 1mM MnCl 2 , 10mM DTT, further wash the column with 3 column volumes of 0.5M NaCl, 50mM Tris pH 8.0, 1mM DTT, 1mMMnCl in a single step 2 elute.
[0153] Appropriate fractions were collected and protein...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Protein concentration | aaaaa | aaaaa |
| Molecular weight | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com